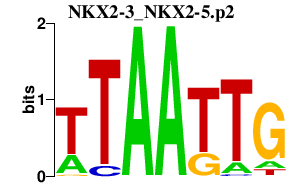
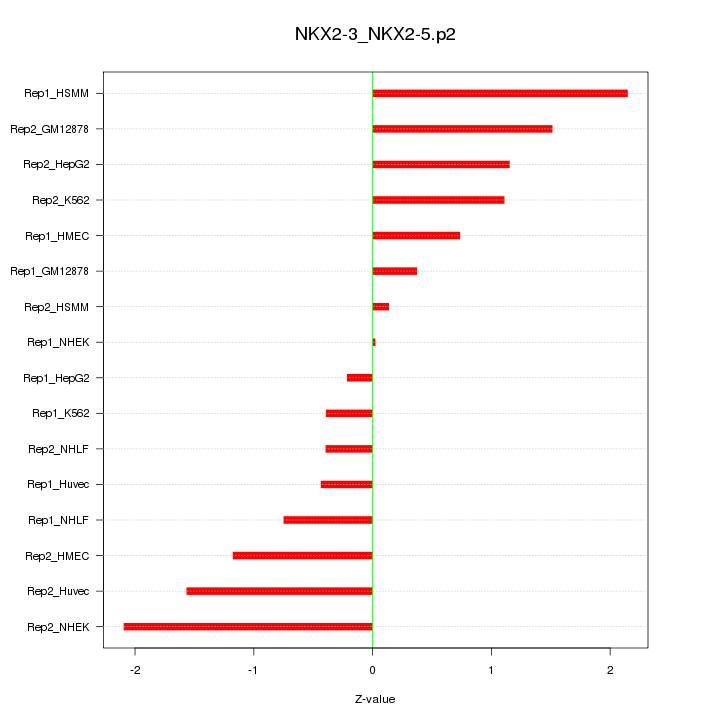
|
chr7_-_13992351
|
3.565
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr17_-_71285992
|
2.629
|
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr9_+_117955890
|
2.014
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr12_-_120725210
|
1.876
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr7_-_27171673
|
1.802
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr7_-_27171648
|
1.723
|
|
HOXA9
|
homeobox A9
|
|
chr7_-_27171600
|
1.647
|
|
HOXA9
|
homeobox A9
|
|
chr12_+_52696908
|
1.625
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr6_+_50789215
|
1.596
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr6_+_155579788
|
1.341
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr20_+_62266217
|
1.256
|
|
MYT1
|
myelin transcription factor 1
|
|
chr4_+_3341521
|
1.241
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr1_+_50342168
|
1.207
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr5_+_175041069
|
1.203
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr5_-_88155360
|
1.197
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr20_+_62266270
|
1.162
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr17_+_7682355
|
1.161
|
|
|
|
|
chr3_+_16191159
|
1.109
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr20_-_62052922
|
1.094
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr22_+_38903825
|
1.090
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_-_127810087
|
1.086
|
NM_001039783
|
TXNRD3IT1
|
thioredoxin reductase 3 intronic transcript 1
|
|
chr1_-_149681265
|
1.082
|
NM_207171
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr19_-_46504831
|
1.008
|
|
|
|
|
chr12_-_28016182
|
0.995
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr1_+_556968
|
0.991
|
|
|
|
|
chr17_-_36209672
|
0.987
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr22_-_36836620
|
0.984
|
NM_025045
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr13_-_93929719
|
0.964
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr2_+_171280046
|
0.957
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr16_+_10878539
|
0.948
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr6_-_33862690
|
0.890
|
NM_001143944
|
LEMD2
|
LEM domain containing 2
|
|
chr12_+_52697160
|
0.876
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr6_-_46401174
|
0.875
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr9_+_129900637
|
0.875
|
NM_052901
NM_001006643
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr2_+_171280182
|
0.873
|
|
|
|
|
chr9_+_129900590
|
0.869
|
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr4_-_38710376
|
0.863
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr14_+_91858677
|
0.849
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr22_-_36836478
|
0.815
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr12_-_51233001
|
0.803
|
NM_033448
|
KRT71
|
keratin 71
|
|
chr13_-_35327798
|
0.797
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr14_-_100105883
|
0.786
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr3_+_46894239
|
0.776
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr15_-_64108718
|
0.773
|
|
|
|
|
chr6_+_16251260
|
0.767
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr1_+_203503214
|
0.748
|
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr17_-_1211263
|
0.741
|
|
YWHAE
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide
|
|
chr17_-_76064842
|
0.736
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr4_+_140806536
|
0.721
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr3_-_152517204
|
0.706
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr5_+_169713458
|
0.693
|
NM_001034838
|
KCNIP1
|
Kv channel interacting protein 1
|
|
chr13_+_97856148
|
0.685
|
|
|
|
|
chr17_-_19290745
|
0.683
|
|
|
|
|
chr16_+_7322751
|
0.678
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr3_+_69895651
|
0.676
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr12_-_89922888
|
0.673
|
NM_004950
|
EPYC
|
epiphycan
|
|
chrX_-_18600108
|
0.670
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr20_+_9146036
|
0.669
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr4_-_83566045
|
0.655
|
|
|
|
|
chr12_-_10173947
|
0.651
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr20_-_43163021
|
0.645
|
NM_002251
|
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1
|
|
chr11_+_7574992
|
0.644
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr2_+_161980865
|
0.643
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr17_+_56843893
|
0.637
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr1_-_149004852
|
0.633
|
|
CTSS
|
cathepsin S
|
|
chr4_+_9094611
|
0.633
|
NM_001040071
|
LOC650293
|
seven transmembrane helix receptor
|
|
chr4_-_153493133
|
0.630
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr22_-_43987010
|
0.627
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr6_+_134800533
|
0.626
|
|
LOC154092
|
hypothetical LOC154092
|
|
chr1_+_17817397
|
0.622
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr14_+_31868229
|
0.614
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr7_-_115458047
|
0.609
|
|
TFEC
|
transcription factor EC
|
|
chr16_-_21571463
|
0.609
|
NM_005849
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chrX_-_19598912
|
0.600
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr13_+_26896614
|
0.600
|
NM_002097
|
GTF3A
|
general transcription factor IIIA
|
|
chr4_-_83566246
|
0.588
|
|
HNRPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr11_+_91724909
|
0.583
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr16_-_21571409
|
0.582
|
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr12_-_51200347
|
0.582
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr7_+_113842284
|
0.573
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr19_-_13925192
|
0.571
|
NM_001146254
NM_001146255
|
PODNL1
|
podocan-like 1
|
|
chr8_-_102029997
|
0.569
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr20_+_31713750
|
0.567
|
NM_080825
|
C20orf144
|
chromosome 20 open reading frame 144
|
|
chr1_+_166516901
|
0.565
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr10_-_32707549
|
0.561
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr9_+_130909926
|
0.545
|
|
|
|
|
chr6_-_86407875
|
0.536
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr2_+_158559936
|
0.536
|
NM_001135098
|
UPP2
|
uridine phosphorylase 2
|
|
chr6_-_74284522
|
0.533
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr7_-_75794919
|
0.533
|
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr19_+_50100843
|
0.530
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr13_-_37070862
|
0.529
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr2_-_20375497
|
0.527
|
|
PUM2
|
pumilio homolog 2 (Drosophila)
|
|
chr18_+_71051713
|
0.525
|
NM_005786
|
TSHZ1
|
teashirt zinc finger homeobox 1
|
|
chr6_+_50894397
|
0.525
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr2_-_215953818
|
0.524
|
|
FN1
|
fibronectin 1
|
|
chr7_-_115458010
|
0.523
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr1_-_180624380
|
0.522
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr20_+_31287462
|
0.517
|
NM_016583
NM_130852
|
PLUNC
|
palate, lung and nasal epithelium associated
|
|
chrX_+_70682204
|
0.517
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr10_+_44726635
|
0.514
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr6_+_12824873
|
0.508
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr17_-_36346240
|
0.507
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr6_+_12825022
|
0.502
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr2_+_173394560
|
0.496
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr12_-_51173238
|
0.495
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr6_-_2580296
|
0.487
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr4_+_71713324
|
0.485
|
NM_031889
|
ENAM
|
enamelin
|
|
chr17_+_72065440
|
0.482
|
|
LOC100507246
|
hypothetical LOC100507246
|
|
chr10_+_80743769
|
0.481
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr7_-_41709191
|
0.478
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr11_+_20001130
|
0.462
|
NM_001111019
|
NAV2
|
neuron navigator 2
|
|
chr10_+_124660206
|
0.461
|
NM_001029888
|
FAM24A
|
family with sequence similarity 24, member A
|
|
chr1_+_149299490
|
0.456
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr10_+_71833920
|
0.455
|
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr7_+_141054621
|
0.453
|
NM_001105558
|
WEE2
|
WEE1 homolog 2 (S. pombe)
|
|
chr1_+_938709
|
0.449
|
NM_005101
|
ISG15
|
ISG15 ubiquitin-like modifier
|
|
chr9_-_205892
|
0.447
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr15_+_91244563
|
0.446
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr18_-_13905534
|
0.445
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chr6_-_169395847
|
0.444
|
|
THBS2
|
thrombospondin 2
|
|
chr17_+_72982919
|
0.437
|
NM_001113495
|
SEPT9
|
septin 9
|
|
chr8_-_17624009
|
0.436
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr9_-_133574928
|
0.435
|
NM_198679
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr20_-_49747397
|
0.434
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr1_-_53972464
|
0.433
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr15_+_97990705
|
0.431
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr1_+_154449389
|
0.431
|
NM_007221
|
PMF1-BGLAP
PMF1
|
PMF1-BGLAP read-through transcript
polyamine-modulated factor 1
|
|
chrX_+_21960841
|
0.430
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chr2_-_190635566
|
0.429
|
NM_005259
|
MSTN
|
myostatin
|
|
chr8_-_72436519
|
0.428
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr6_-_52882419
|
0.423
|
NM_000847
|
GSTA3
|
glutathione S-transferase alpha 3
|
|
chr22_-_16127520
|
0.421
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr7_-_104696678
|
0.419
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr2_+_161981138
|
0.417
|
|
TBR1
|
T-box, brain, 1
|
|
chr4_+_155703581
|
0.415
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr16_-_20246309
|
0.415
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr8_+_55691179
|
0.413
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr10_+_71833905
|
0.410
|
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr11_-_2108035
|
0.409
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_+_54587882
|
0.408
|
|
PRR11
|
proline rich 11
|
|
chr1_+_208568218
|
0.407
|
NM_001122834
NM_001170564
|
HHAT
|
hedgehog acyltransferase
|
|
chr1_+_149299530
|
0.406
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr6_+_37081387
|
0.405
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr11_-_46364681
|
0.404
|
NM_000741
|
CHRM4
|
cholinergic receptor, muscarinic 4
|
|
chr3_-_55489973
|
0.404
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr2_+_161981046
|
0.402
|
|
TBR1
|
T-box, brain, 1
|
|
chr5_+_174084137
|
0.402
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr12_-_9913199
|
0.402
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr16_+_20682812
|
0.398
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr1_+_559129
|
0.397
|
|
|
|
|
chr20_-_7869068
|
0.395
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr7_+_129771865
|
0.393
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr4_-_176970895
|
0.393
|
|
GPM6A
|
glycoprotein M6A
|
|
chr17_-_10393664
|
0.391
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr20_-_1921451
|
0.390
|
NM_001190892
|
PDYN
|
prodynorphin
|
|
chr12_-_88269884
|
0.386
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr1_+_149299504
|
0.386
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr6_-_46401363
|
0.385
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chrX_+_68752635
|
0.383
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr2_-_178681311
|
0.382
|
NM_001077197
|
PDE11A
|
phosphodiesterase 11A
|
|
chr11_+_65023218
|
0.380
|
|
|
|
|
chr8_-_122722674
|
0.380
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr14_+_99633570
|
0.379
|
|
EVL
|
Enah/Vasp-like
|
|
chr10_+_111755715
|
0.378
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr11_+_128067598
|
0.378
|
NM_001167681
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr4_+_71418886
|
0.373
|
NM_212557
|
AMTN
|
amelotin
|
|
chr6_+_158653679
|
0.372
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chrX_+_90976314
|
0.372
|
NM_014522
NM_032968
NM_032969
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr6_+_152168500
|
0.371
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|
|
chr4_+_71142893
|
0.368
|
NM_005212
|
CSN3
|
casein kappa
|
|
chr3_-_188935363
|
0.366
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr16_+_79829794
|
0.366
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr3_+_160269836
|
0.363
|
|
IQCJ
|
IQ motif containing J
|
|
chr3_+_8750485
|
0.362
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr20_+_43691797
|
0.361
|
NM_080753
|
WFDC10A
|
WAP four-disulfide core domain 10A
|
|
chr12_+_12829807
|
0.361
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr12_+_48443232
|
0.359
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr5_+_71538456
|
0.359
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr5_+_1278469
|
0.357
|
NM_182632
|
SLC6A18
|
solute carrier family 6, member 18
|
|
chr17_-_64649608
|
0.356
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr3_-_11585264
|
0.354
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr20_+_19863716
|
0.350
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr11_+_65026382
|
0.350
|
|
|
|
|
chr17_+_38412267
|
0.350
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr10_-_105835609
|
0.347
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr3_+_160269734
|
0.347
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr2_+_220017617
|
0.346
|
|
SPEG
|
SPEG complex locus
|
|
chr1_-_180624494
|
0.344
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr10_-_97190885
|
0.343
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr20_-_21442663
|
0.343
|
NM_002509
|
NKX2-2
|
NK2 homeobox 2
|
|
chr1_-_109957195
|
0.342
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr2_-_160764819
|
0.340
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr10_+_71833866
|
0.340
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr9_-_135379888
|
0.337
|
NM_001080483
|
TMEM8C
|
transmembrane protein 8C
|
|
chr2_+_157822355
|
0.336
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr1_+_557269
|
0.336
|
|
|
|
|
chr10_-_64246129
|
0.334
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr6_-_169395972
|
0.333
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr3_-_139960874
|
0.332
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|