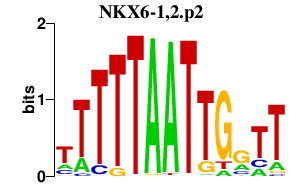
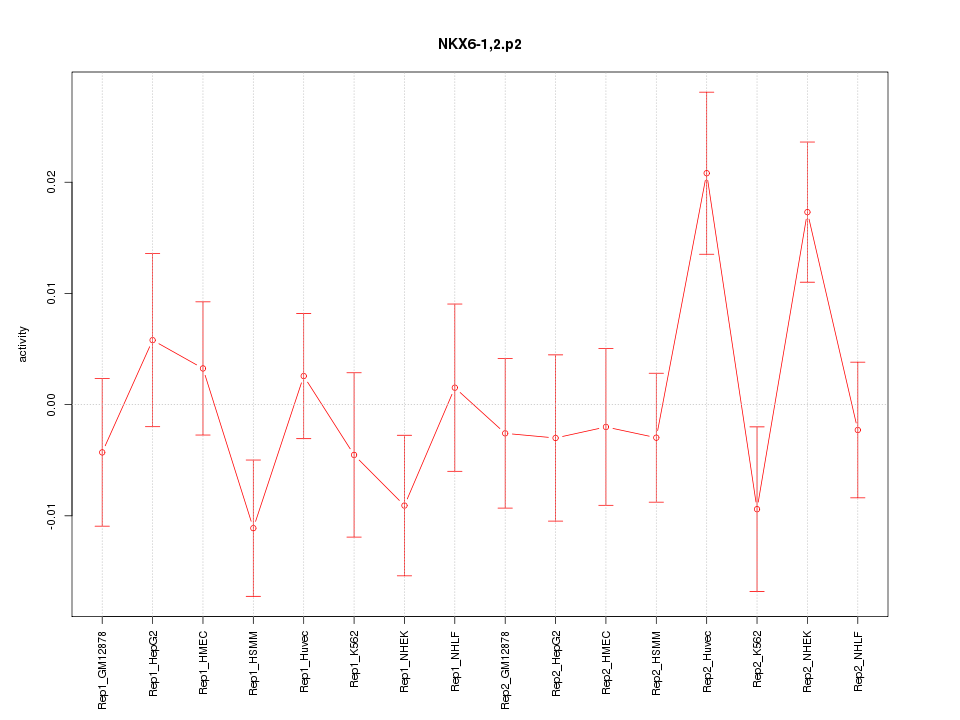
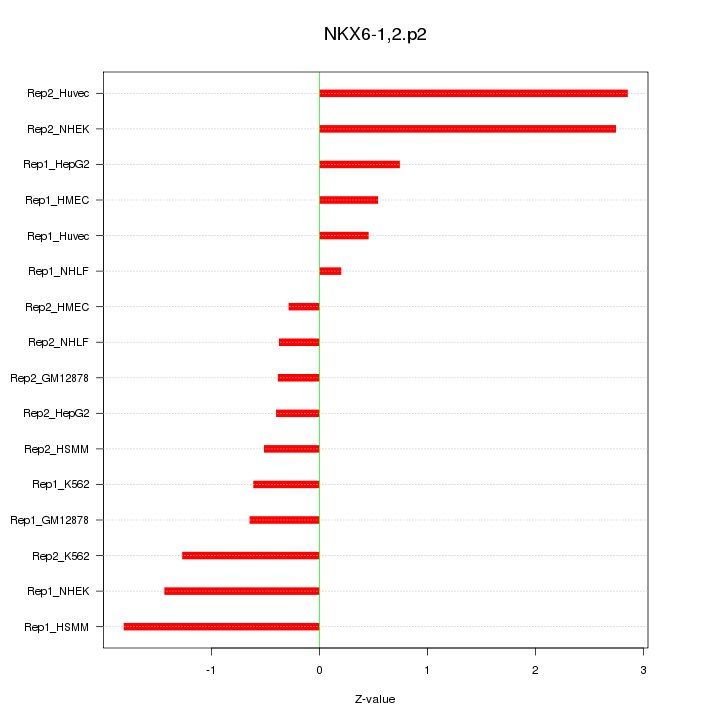
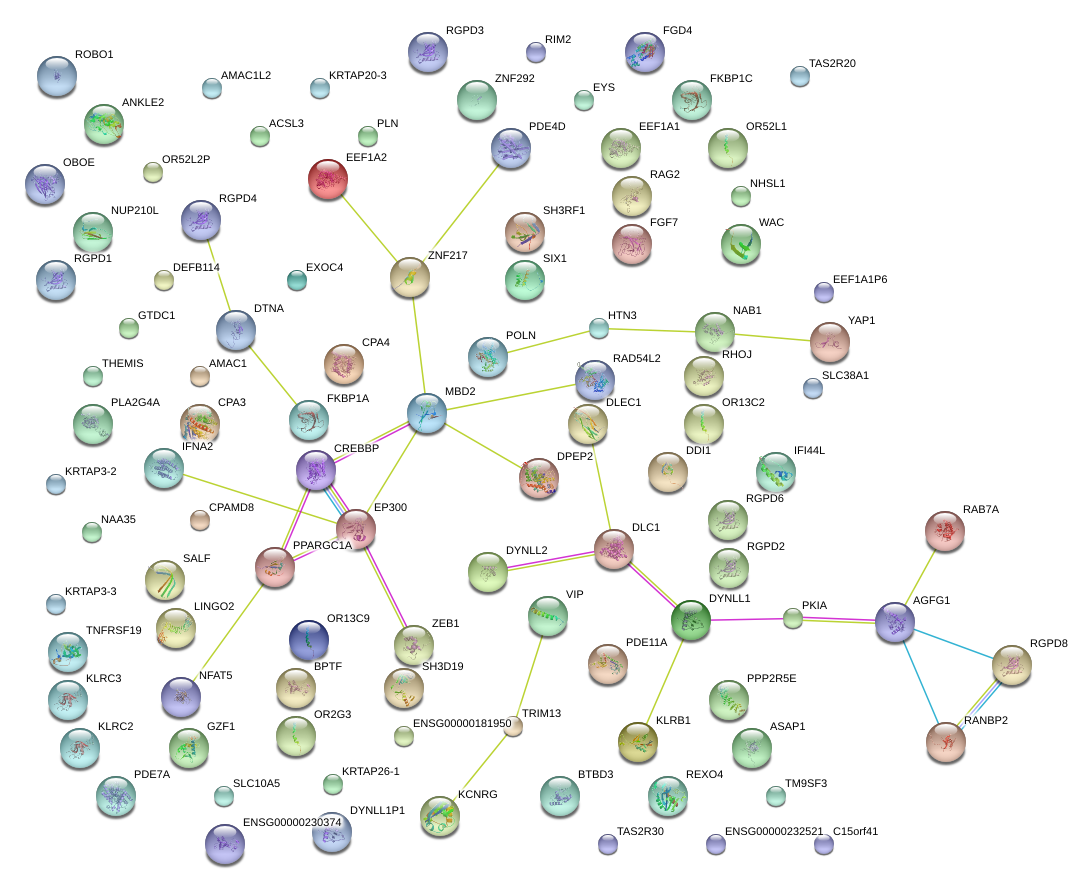
|
chr9_-_21375258
|
2.631
|
NM_000605
|
IFNA2
|
interferon, alpha 2
|
|
chr5_-_58331515
|
2.270
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_79899748
|
1.861
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr14_+_62740878
|
1.638
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr13_+_23042722
|
1.516
|
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr6_-_128263868
|
1.452
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr4_+_70928718
|
1.204
|
NM_000200
|
HTN3
|
histatin 3
|
|
chr8_-_82769761
|
1.181
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr15_+_34720097
|
1.147
|
NM_032499
|
C15orf41
|
chromosome 15 open reading frame 41
|
|
chr20_+_23293020
|
1.115
|
NM_022482
|
GZF1
|
GDNF-inducible zinc finger protein 1
|
|
chr2_+_191222077
|
1.051
|
NM_005966
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1)
|
|
chr12_+_32546243
|
1.044
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr3_+_130014957
|
1.033
|
|
RAB7A
|
RAB7A, member RAS oncogene family
|
|
chr3_-_121065112
|
0.995
|
|
|
|
|
chr1_+_245835478
|
0.991
|
NM_001001914
|
OR2G3
|
olfactory receptor, family 2, subfamily G, member 3
|
|
chr6_-_66346417
|
0.988
|
NM_198283
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr14_-_63076155
|
0.975
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr20_+_11846536
|
0.966
|
NM_014962
|
BTBD3
|
BTB (POZ) domain containing 3
|
|
chr12_-_131816604
|
0.958
|
|
ANKLE2
|
ankyrin repeat and LEM domain containing 2
|
|
chr6_+_118976134
|
0.956
|
NM_002667
|
PLN
|
phospholamban
|
|
chr2_+_228064531
|
0.920
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr20_-_51633042
|
0.920
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr12_-_44919818
|
0.913
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr8_-_13018123
|
0.890
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr11_-_5964790
|
0.889
|
NM_001005173
|
OR52L1
|
olfactory receptor, family 52, subfamily L, member 1
|
|
chr6_+_88026338
|
0.878
|
|
ZNF292
|
zinc finger protein 292
|
|
chr6_-_138935359
|
0.850
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr15_+_47502666
|
0.842
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr1_+_185064654
|
0.839
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr2_+_108751033
|
0.832
|
|
RANBP2
|
RAN binding protein 2
|
|
chr18_-_49944986
|
0.822
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr17_+_55269974
|
0.820
|
|
|
|
|
chr21_+_30937053
|
0.810
|
NM_001128077
|
KRTAP20-3
|
keratin associated protein 20-3
|
|
chr2_-_178495768
|
0.803
|
NM_001077358
|
PDE11A
|
phosphodiesterase 11A
|
|
chr2_-_144711375
|
0.795
|
NM_024659
|
GTDC1
|
glycosyltransferase-like domain containing 1
|
|
chr6_-_138862271
|
0.769
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr8_-_66863841
|
0.768
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr6_+_153113624
|
0.764
|
NM_003381
NM_194435
|
VIP
|
vasoactive intestinal peptide
|
|
chr9_-_28709302
|
0.751
|
NM_152570
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr8_+_11225904
|
0.738
|
NM_054028
|
AMAC1L2
|
acyl-malonyl condensing enzyme 1-like 2
|
|
chr12_-_11106159
|
0.737
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr11_+_101488380
|
0.735
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr1_+_78858675
|
0.731
|
NM_006820
|
IFI44L
|
interferon-induced protein 44-like
|
|
chr8_+_79591074
|
0.706
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr9_+_87745876
|
0.705
|
NM_024635
|
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit
|
|
chr1_-_152394215
|
0.699
|
NM_001159484
NM_207308
|
NUP210L
|
nucleoporin 210kDa-like
|
|
chr4_-_170314382
|
0.696
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr3_+_150065732
|
0.689
|
NM_001870
|
CPA3
|
carboxypeptidase A3 (mast cell)
|
|
chr14_-_60185659
|
0.679
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr6_-_50039776
|
0.671
|
NM_001037499
|
DEFB114
|
defensin, beta 114
|
|
chr20_-_1298493
|
0.646
|
|
FKBP1A
|
FK506 binding protein 1A, 12kDa
|
|
chr8_-_131483395
|
0.643
|
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr8_+_104900591
|
0.643
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr21_-_30614477
|
0.629
|
NM_203405
|
KRTAP26-1
|
keratin associated protein 26-1
|
|
chr7_+_132588359
|
0.627
|
NM_001037126
NM_021807
|
EXOC4
|
exocyst complex component 4
|
|
chr11_+_103412517
|
0.623
|
NM_001001711
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae)
|
|
chr12_-_10464308
|
0.619
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr12_-_11041740
|
0.614
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr17_+_63367052
|
0.607
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr10_+_28918672
|
0.596
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr6_-_138862273
|
0.596
|
|
|
|
|
chr10_-_98311813
|
0.595
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr4_-_23500706
|
0.593
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr9_-_106407728
|
0.589
|
NM_001004481
|
OR13C2
|
olfactory receptor, family 13, subfamily C, member 2
|
|
chr18_+_30544193
|
0.588
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr13_+_49487390
|
0.585
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr10_+_31650069
|
0.577
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr11_-_36576352
|
0.567
|
NM_000536
|
RAG2
|
recombination activating gene 2
|
|
chr16_+_68238460
|
0.563
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr16_-_3783475
|
0.561
|
|
CREBBP
|
CREB binding protein
|
|
chr17_-_36403910
|
0.559
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr2_+_223433919
|
0.555
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr4_-_152368492
|
0.555
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr22_+_29696662
|
0.554
|
|
|
|
|
chr4_-_2200755
|
0.552
|
NM_181808
|
POLN
|
polymerase (DNA directed) nu
|
|
chr3_+_51550635
|
0.550
|
NM_015106
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae)
|
|
chr7_-_22518200
|
0.550
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr12_+_9871343
|
0.544
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr20_+_16676997
|
0.537
|
NM_020157
|
OTOR
|
otoraplin
|
|
chr1_-_115071195
|
0.532
|
|
CSDE1
|
cold shock domain containing E1, RNA-binding
|
|
chr13_+_51496679
|
0.531
|
NM_021645
|
ALG11
UTP14C
|
asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase homolog (yeast)
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast)
|
|
chr17_+_58625086
|
0.530
|
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr8_+_24354387
|
0.527
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chrX_-_102973762
|
0.525
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr6_+_132901119
|
0.524
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr12_-_44644216
|
0.517
|
|
|
|
|
chr2_+_108360798
|
0.514
|
NM_006588
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4
|
|
chr10_+_50557689
|
0.513
|
NM_001042427
NM_182554
|
C10orf53
|
chromosome 10 open reading frame 53
|
|
chr3_-_194579207
|
0.511
|
NM_198505
|
ATP13A5
|
ATPase type 13A5
|
|
chr11_-_7684516
|
0.506
|
NM_198185
|
OVCH2
|
ovochymase 2 (gene/pseudogene)
|
|
chr11_+_93916653
|
0.496
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chr13_+_96726415
|
0.494
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr14_-_94669547
|
0.488
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr1_+_144188375
|
0.488
|
NM_153713
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr6_-_132064233
|
0.487
|
NM_030908
|
OR2A4
|
olfactory receptor, family 2, subfamily A, member 4
|
|
chr3_-_25654659
|
0.482
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr13_-_45323757
|
0.480
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr15_+_32048380
|
0.476
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr10_+_124135574
|
0.474
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr11_-_57964221
|
0.474
|
NM_001004733
|
OR5B12
|
olfactory receptor, family 5, subfamily B, member 12
|
|
chr4_-_17415918
|
0.470
|
|
DCAF16
|
DDB1 and CUL4 associated factor 16
|
|
chr1_-_246857051
|
0.470
|
NM_001001964
|
OR2T11
|
olfactory receptor, family 2, subfamily T, member 11
|
|
chr19_-_53912025
|
0.468
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr1_+_115198977
|
0.468
|
NM_003176
|
SYCP1
|
synaptonemal complex protein 1
|
|
chr14_-_23727826
|
0.467
|
|
IPO4
|
importin 4
|
|
chr5_+_166644420
|
0.467
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr2_-_183095308
|
0.458
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr8_+_79590890
|
0.453
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr3_+_99592199
|
0.452
|
NM_001005516
|
OR5K3
|
olfactory receptor, family 5, subfamily K, member 3
|
|
chr4_+_53975597
|
0.452
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr4_-_87593306
|
0.440
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr17_-_36376663
|
0.438
|
NM_213656
|
KRT39
|
keratin 39
|
|
chrX_-_34060348
|
0.434
|
NM_203408
|
FAM47A
|
family with sequence similarity 47, member A
|
|
chr14_-_23727850
|
0.430
|
NM_024658
|
IPO4
|
importin 4
|
|
chr19_+_60962318
|
0.428
|
NM_001145014
|
RFPL4A
|
ret finger protein-like 4A
|
|
chr8_-_91164100
|
0.427
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr11_-_55815113
|
0.427
|
NM_001005199
|
OR8H1
|
olfactory receptor, family 8, subfamily H, member 1
|
|
chr13_-_102517196
|
0.426
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr2_-_65447267
|
0.425
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr4_-_146315341
|
0.414
|
NM_017493
|
OTUD4
|
OTU domain containing 4
|
|
chr3_+_175641340
|
0.412
|
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chrX_+_102727068
|
0.405
|
NM_001006935
NM_001006937
NM_024863
|
TCEAL4
|
transcription elongation factor A (SII)-like 4
|
|
chr18_+_59705918
|
0.404
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr15_-_74091817
|
0.403
|
NM_138573
|
NRG4
|
neuregulin 4
|
|
chr4_-_90978469
|
0.403
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr2_+_183316574
|
0.401
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr2_-_165768798
|
0.401
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr9_+_115303527
|
0.401
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_-_65447370
|
0.394
|
NM_001128210
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr6_-_87861492
|
0.393
|
NM_000735
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr4_-_70760381
|
0.392
|
NM_005420
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1
|
|
chr5_+_161427225
|
0.391
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr2_+_118288724
|
0.391
|
NM_006773
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr2_-_210876560
|
0.389
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr9_-_21340885
|
0.387
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr2_+_118288754
|
0.385
|
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr2_-_121964373
|
0.385
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr2_-_210726539
|
0.384
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr3_+_45902999
|
0.384
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr2_+_118288753
|
0.383
|
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18
|
|
chr10_-_69661860
|
0.383
|
NM_145178
|
ATOH7
|
atonal homolog 7 (Drosophila)
|
|
chr8_-_52884456
|
0.383
|
NM_144651
|
PXDNL
|
peroxidasin homolog (Drosophila)-like
|
|
chr8_-_82522175
|
0.381
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr17_+_36228869
|
0.381
|
NM_001195387
NM_145274
NM_001195386
|
TMEM99
|
transmembrane protein 99
|
|
chr15_-_74941339
|
0.379
|
NM_001145923
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr11_-_27637771
|
0.375
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_57883117
|
0.374
|
NM_001005489
|
OR5B17
|
olfactory receptor, family 5, subfamily B, member 17
|
|
chr9_-_72673753
|
0.371
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr7_+_72034735
|
0.370
|
|
POM121
|
POM121 membrane glycoprotein
|
|
chr12_+_63090008
|
0.366
|
|
XPOT
|
exportin, tRNA (nuclear export receptor for tRNAs)
|
|
chr1_+_84402962
|
0.366
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr20_+_56455866
|
0.366
|
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr7_-_122627235
|
0.364
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr10_+_86121526
|
0.360
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr2_+_224530361
|
0.360
|
NM_022915
|
MRPL44
|
mitochondrial ribosomal protein L44
|
|
chr7_-_26198673
|
0.359
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr12_-_53664722
|
0.356
|
NM_001136030
|
KIAA0748
|
KIAA0748
|
|
chr3_-_197726587
|
0.355
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr14_-_56342097
|
0.355
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr9_-_13240313
|
0.348
|
NM_003829
|
MPDZ
|
multiple PDZ domain protein
|
|
chrX_-_138114100
|
0.348
|
|
FGF13
|
fibroblast growth factor 13
|
|
chr2_-_37397669
|
0.347
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr4_+_22608252
|
0.346
|
|
|
|
|
chr6_+_53991672
|
0.345
|
NM_138569
|
C6orf142
|
chromosome 6 open reading frame 142
|
|
chr7_-_107667728
|
0.344
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr21_-_30786145
|
0.342
|
NM_181609
|
KRTAP19-3
|
keratin associated protein 19-3
|
|
chr3_-_166397142
|
0.341
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr21_-_31332665
|
0.341
|
NM_001099219
|
KRTAP19-8
|
keratin associated protein 19-8
|
|
chr3_+_99484421
|
0.338
|
NM_001005482
|
OR5H2
|
olfactory receptor, family 5, subfamily H, member 2
|
|
chr2_+_137464931
|
0.337
|
NM_001080427
|
THSD7B
|
thrombospondin, type I, domain containing 7B
|
|
chr19_+_12036513
|
0.335
|
NM_001136501
|
ZNF844
|
zinc finger protein 844
|
|
chr20_-_18425811
|
0.334
|
NM_006606
|
RBBP9
|
retinoblastoma binding protein 9
|
|
chr5_-_43326922
|
0.334
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr18_-_29882555
|
0.333
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr14_+_59628381
|
0.333
|
NM_022495
|
C14orf135
|
chromosome 14 open reading frame 135
|
|
chr1_+_85300580
|
0.333
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr12_-_11075272
|
0.332
|
NM_176885
|
TAS2R31
|
taste receptor, type 2, member 31
|
|
chrX_-_76824726
|
0.330
|
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr12_+_49155798
|
0.330
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr11_+_55799604
|
0.330
|
NM_001004745
|
OR5T1
|
olfactory receptor, family 5, subfamily T, member 1
|
|
chr9_-_106420305
|
0.329
|
NM_001001956
|
OR13C9
|
olfactory receptor, family 13, subfamily C, member 9
|
|
chr7_+_142053693
|
0.327
|
NM_001190487
|
MTRNR2L6
|
MT-RNR2-like 6
|
|
chr3_+_12367993
|
0.327
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr8_+_133060353
|
0.326
|
|
EFR3A
|
EFR3 homolog A (S. cerevisiae)
|
|
chr1_+_103898844
|
0.325
|
NM_020978
|
AMY2B
|
amylase, alpha 2B (pancreatic)
|
|
chr21_-_30781537
|
0.325
|
NM_181608
|
KRTAP19-2
|
keratin associated protein 19-2
|
|
chr1_-_245988330
|
0.325
|
NM_012353
|
OR1C1
|
olfactory receptor, family 1, subfamily C, member 1
|
|
chr2_+_37282516
|
0.324
|
|
LOC100505876
|
hypothetical LOC100505876
|
|
chr17_+_36636404
|
0.322
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr5_-_150707281
|
0.320
|
NM_181776
|
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2
|
|
chr3_+_39346200
|
0.319
|
NM_005201
|
CCR8
|
chemokine (C-C motif) receptor 8
|
|
chr4_+_71283383
|
0.318
|
NM_006685
|
SMR3B
|
submaxillary gland androgen regulated protein 3B
|
|
chr3_-_51912376
|
0.316
|
NM_152397
|
IQCF1
|
IQ motif containing F1
|
|
chr3_+_63859114
|
0.316
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr2_-_37150876
|
0.316
|
|
HEATR5B
|
HEAT repeat containing 5B
|
|
chr10_-_33264491
|
0.314
|
NM_033668
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr14_-_19867372
|
0.314
|
NM_182852
NM_182851
|
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase
|
|
chr1_+_61642287
|
0.313
|
|
NFIA
|
nuclear factor I/A
|
|
chr5_+_161207517
|
0.313
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chrX_+_85404626
|
0.313
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|