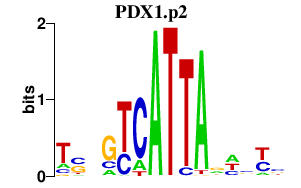
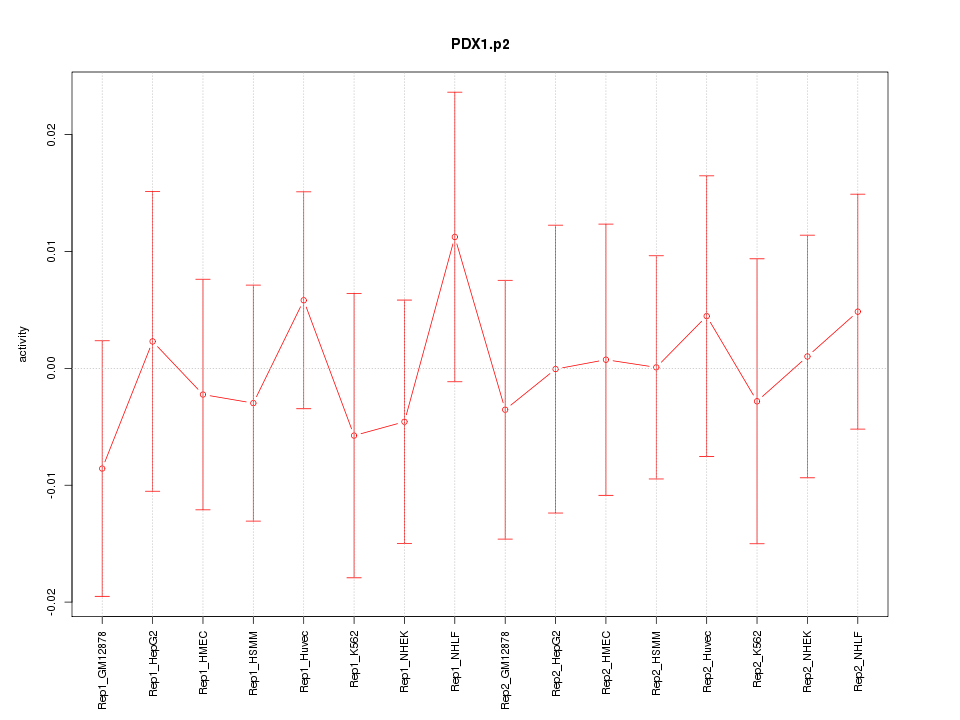
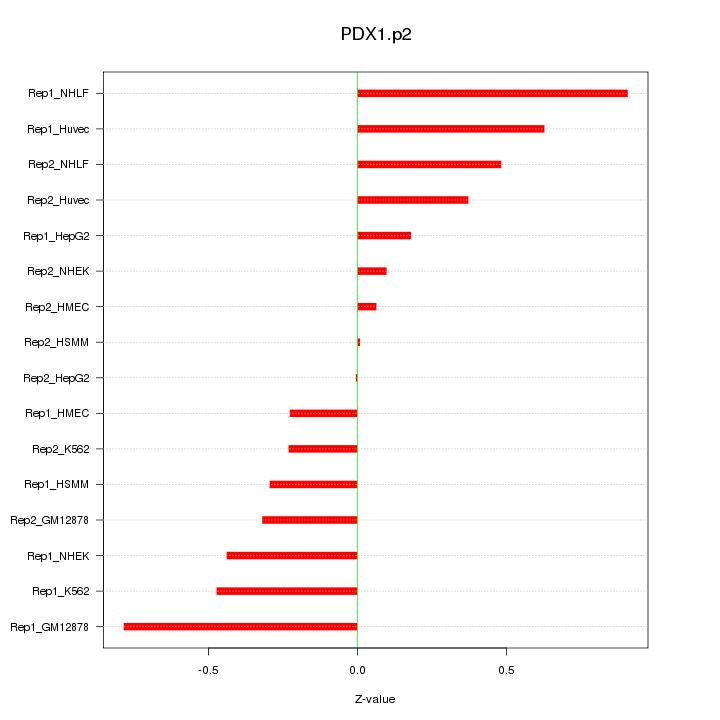
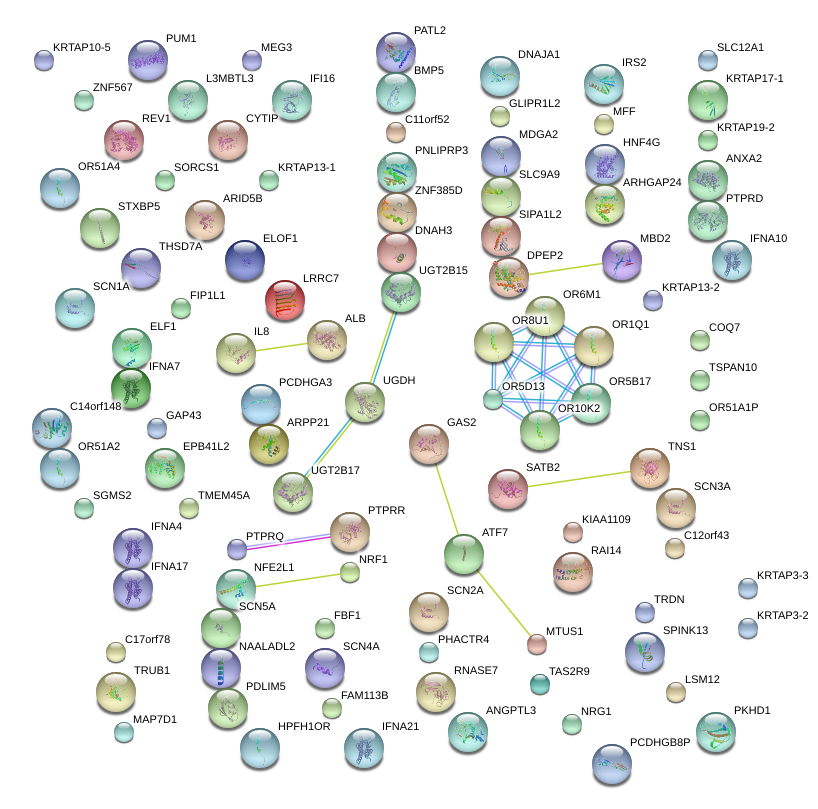
|
chr3_-_21767819
|
0.788
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr10_+_63478974
|
0.768
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr1_+_62835759
|
0.614
|
NM_014495
|
ANGPTL3
|
angiopoietin-like 3
|
|
chr15_+_46285789
|
0.517
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr11_+_55899675
|
0.450
|
NM_001005204
NM_001013356
|
OR8U1
OR8U8
|
olfactory receptor, family 8, subfamily U, member 1
olfactory receptor, family 8, subfamily U, member 8
|
|
chr2_-_158008763
|
0.408
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr4_+_109033868
|
0.399
|
NM_001136257
NM_152621
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr3_+_101720753
|
0.398
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr11_+_22652957
|
0.294
|
|
GAS2
|
growth arrest-specific 2
|
|
chr4_+_86615275
|
0.282
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr6_+_130381408
|
0.273
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr21_-_30781537
|
0.256
|
NM_181608
|
KRTAP19-2
|
keratin associated protein 19-2
|
|
chr11_-_123182266
|
0.252
|
NM_001005325
|
OR6M1
|
olfactory receptor, family 6, subfamily M, member 1
|
|
chr11_+_22652894
|
0.219
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr2_-_165768798
|
0.213
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr5_+_34720368
|
0.204
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr19_+_41872141
|
0.195
|
NM_152603
|
ZNF567
|
zinc finger protein 567
|
|
chr4_-_69116839
|
0.190
|
NM_001077
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chr1_+_157246305
|
0.181
|
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr5_+_147628567
|
0.174
|
NM_001040129
|
SPINK13
|
serine peptidase inhibitor, Kazal type 13 (putative)
|
|
chr17_-_36725470
|
0.170
|
NM_031964
|
KRTAP17-1
|
keratin associated protein 17-1
|
|
chr15_-_42756377
|
0.162
|
NM_001145112
|
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast)
|
|
chr8_+_76614757
|
0.153
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr4_+_95595419
|
0.150
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr9_+_33016482
|
0.150
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr1_+_36409277
|
0.142
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr9_+_33614214
|
0.136
|
|
ANXA2P2
|
annexin A2 pseudogene 2
|
|
chr9_-_8723893
|
0.132
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr10_+_118177371
|
0.129
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr11_-_5130174
|
0.121
|
NM_012375
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1
|
|
chr4_+_123380569
|
0.120
|
|
KIAA1109
|
KIAA1109
|
|
chr1_-_156657279
|
0.119
|
NM_001004476
|
OR10K2
|
olfactory receptor, family 10, subfamily K, member 2
|
|
chr11_-_4924905
|
0.119
|
NM_001005329
|
OR51A4
|
olfactory receptor, family 51, subfamily A, member 4
|
|
chr8_-_17624304
|
0.119
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr6_-_55848346
|
0.119
|
|
BMP5
|
bone morphogenetic protein 5
|
|
chr16_-_21078262
|
0.116
|
NM_017539
|
DNAH3
|
dynein, axonemal, heavy chain 3
|
|
chr17_-_36409632
|
0.114
|
NM_031959
|
KRTAP3-2
|
keratin associated protein 3-2
|
|
chr14_-_46882144
|
0.114
|
NM_182830
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr9_+_124416768
|
0.111
|
NM_012364
|
OR1Q1
|
olfactory receptor, family 1, subfamily Q, member 1
|
|
chr12_-_52306348
|
0.109
|
NM_001130060
NM_006856
|
ATF7
|
activating transcription factor 7
|
|
chr1_+_28637247
|
0.109
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr16_+_18986417
|
0.108
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr12_-_52305900
|
0.105
|
|
ATF7
|
activating transcription factor 7
|
|
chr17_-_36403910
|
0.104
|
NM_033185
|
KRTAP3-3
|
keratin associated protein 3-3
|
|
chr13_-_109236897
|
0.103
|
NM_003749
|
IRS2
|
insulin receptor substrate 2
|
|
chr12_-_119938667
|
0.101
|
NM_022895
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr8_+_32698892
|
0.101
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr17_+_7900926
|
0.101
|
|
|
|
|
chr14_+_100365162
|
0.100
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr3_-_145049853
|
0.098
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr6_+_147566559
|
0.097
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr6_-_52060327
|
0.097
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr4_+_74500748
|
0.096
|
|
ALB
|
albumin
|
|
chr1_+_69998445
|
0.094
|
NM_020794
|
LRRC7
|
leucine rich repeat containing 7
|
|
chr6_-_131333173
|
0.091
|
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr18_-_49944986
|
0.088
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr13_-_40491491
|
0.088
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr10_+_116687904
|
0.087
|
NM_139169
|
TRUB1
|
TruB pseudouridine (psi) synthase homolog 1 (E. coli)
|
|
chr21_-_44824907
|
0.086
|
NM_198694
|
KRTAP10-5
|
keratin associated protein 10-5
|
|
chr3_+_35697432
|
0.086
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr14_+_20580224
|
0.083
|
NM_032572
|
RNASE7
|
ribonuclease, RNase A family, 7
|
|
chr2_-_218516950
|
0.082
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr2_+_227900471
|
0.081
|
NM_020194
|
MFF
|
mitochondrial fission factor
|
|
chr12_-_119938593
|
0.081
|
|
C12orf43
|
chromosome 12 open reading frame 43
|
|
chr7_+_129057152
|
0.081
|
NM_001040110
|
NRF1
|
nuclear respiratory factor 1
|
|
chr3_+_116824840
|
0.080
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr6_-_55848317
|
0.078
|
NM_021073
|
BMP5
|
bone morphogenetic protein 5
|
|
chr4_+_53975597
|
0.076
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr14_-_76958984
|
0.076
|
NM_138791
NM_001113475
|
C14orf148
|
chromosome 14 open reading frame 148
|
|
chr3_+_176059763
|
0.075
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr16_+_18986459
|
0.075
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr21_-_30666417
|
0.074
|
NM_181621
|
KRTAP13-2
|
keratin associated protein 13-2
|
|
chr5_+_140698537
|
0.074
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr1_-_230717850
|
0.072
|
NM_020808
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2
|
|
chr17_+_32807040
|
0.071
|
NM_173625
|
C17orf78
|
chromosome 17 open reading frame 78
|
|
chr12_+_74071145
|
0.069
|
NM_152436
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr2_-_199922047
|
0.068
|
|
SATB2
|
SATB homeobox 2
|
|
chr3_+_35658852
|
0.068
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr11_+_111294810
|
0.066
|
NM_080659
|
C11orf52
|
chromosome 11 open reading frame 52
|
|
chr4_+_74825138
|
0.066
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr10_-_108448990
|
0.065
|
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chr7_-_11643113
|
0.065
|
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr12_+_45896511
|
0.065
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr4_-_39199527
|
0.060
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
|
chr12_+_79362256
|
0.059
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr2_-_99412694
|
0.059
|
|
REV1
|
REV1 homolog (S. cerevisiae)
|
|
chr12_-_10853940
|
0.059
|
NM_023917
|
TAS2R9
|
taste receptor, type 2, member 9
|
|
chr1_-_31251315
|
0.058
|
|
PUM1
|
pumilio homolog 1 (Drosophila)
|
|
chr11_-_57883117
|
0.058
|
NM_001005489
|
OR5B17
|
olfactory receptor, family 5, subfamily B, member 17
|
|
chr17_-_39499527
|
0.056
|
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr11_+_55297489
|
0.056
|
NM_001001967
|
OR5D13
|
olfactory receptor, family 5, subfamily D, member 13
|
|
chr6_-_123999640
|
0.056
|
NM_006073
|
TRDN
|
triadin
|
|
chr7_+_116182676
|
0.053
|
|
|
|
|
chr9_-_21218132
|
0.048
|
NM_021268
|
IFNA17
|
interferon, alpha 17
|
|
chr16_-_70339666
|
0.047
|
|
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit
|
|
chr1_-_150328153
|
0.047
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr10_+_90474280
|
0.045
|
NM_001080518
|
LIPK
|
lipase, family member K
|
|
chr17_-_36418869
|
0.043
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr12_-_102968044
|
0.043
|
NM_031302
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr5_+_179934143
|
0.043
|
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6
|
|
chr2_+_185171480
|
0.042
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr16_+_18986721
|
0.042
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr3_+_99350859
|
0.042
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr12_+_10351683
|
0.041
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr1_-_246869181
|
0.041
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr17_-_7023378
|
0.040
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr12_+_74071116
|
0.040
|
|
GLIPR1L2
|
GLI pathogenesis-related 1 like 2
|
|
chr1_-_202402043
|
0.040
|
NM_000537
|
REN
|
renin
|
|
chr10_-_56230953
|
0.039
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr3_-_47909001
|
0.039
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr18_-_28247193
|
0.039
|
|
|
|
|
chr8_+_11877238
|
0.038
|
NM_001033017
|
DEFB135
|
defensin, beta 135
|
|
chr3_-_110155237
|
0.038
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr8_-_20084892
|
0.036
|
NM_001135691
NM_001142324
NM_001142325
NM_003053
|
SLC18A1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr4_+_169250281
|
0.035
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr2_+_203763649
|
0.035
|
|
RPL12
|
ribosomal protein L12
|
|
chr18_+_29573152
|
0.034
|
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr2_-_121964373
|
0.032
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr8_+_26569709
|
0.032
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr17_-_45629401
|
0.032
|
|
|
|
|
chr1_-_161379606
|
0.032
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr7_+_104468614
|
0.031
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr1_-_159786441
|
0.031
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr1_+_161305188
|
0.031
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr2_+_105799354
|
0.031
|
|
NCK2
|
NCK adaptor protein 2
|
|
chr12_+_4569504
|
0.030
|
NM_003845
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4
|
|
chr11_+_123639909
|
0.030
|
NM_001005198
|
OR8G5
|
olfactory receptor, family 8, subfamily G, member 5
|
|
chr7_+_93373755
|
0.029
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chrX_-_131375217
|
0.028
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chrX_+_123307812
|
0.028
|
NM_001114937
NM_002351
|
SH2D1A
|
SH2 domain containing 1A
|
|
chr2_+_169634330
|
0.027
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chrX_+_118592879
|
0.027
|
|
UBE2A
|
ubiquitin-conjugating enzyme E2A (RAD6 homolog)
|
|
chr6_+_144654565
|
0.027
|
NM_007124
|
UTRN
|
utrophin
|
|
chr10_+_126620681
|
0.026
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chr17_-_36444626
|
0.026
|
NM_030966
|
KRTAP1-3
|
keratin associated protein 1-3
|
|
chr7_+_156128431
|
0.026
|
NM_001184997
|
RNF32
|
ring finger protein 32
|
|
chr14_-_57133361
|
0.026
|
NM_001080455
|
SLC35F4
|
solute carrier family 35, member F4
|
|
chr6_-_26151863
|
0.026
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chrX_+_118592568
|
0.026
|
|
UBE2A
|
ubiquitin-conjugating enzyme E2A (RAD6 homolog)
|
|
chr11_+_6823458
|
0.026
|
NM_178168
|
OR10A5
|
olfactory receptor, family 10, subfamily A, member 5
|
|
chr12_+_21417068
|
0.025
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr11_+_88550687
|
0.025
|
NM_000372
|
TYR
|
tyrosinase (oculocutaneous albinism IA)
|
|
chr3_+_149930656
|
0.025
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr4_+_187424336
|
0.024
|
|
F11
|
coagulation factor XI
|
|
chr12_-_101835510
|
0.024
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr10_-_62002672
|
0.024
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chrX_-_62697473
|
0.023
|
|
LOC92249
|
hypothetical LOC92249
|
|
chr12_+_45896318
|
0.022
|
NM_138371
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr2_+_24660849
|
0.022
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr8_+_110443881
|
0.022
|
NM_177531
|
PKHD1L1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1
|
|
chr8_-_7331313
|
0.021
|
NM_152251
NM_001040704
|
DEFB106A
DEFB106B
|
defensin, beta 106A
defensin, beta 106B
|
|
chr2_+_38746742
|
0.021
|
|
GALM
|
galactose mutarotase (aldose 1-epimerase)
|
|
chr11_-_55661769
|
0.021
|
NM_001004064
|
OR8J3
|
olfactory receptor, family 8, subfamily J, member 3
|
|
chr2_+_17861266
|
0.019
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr18_+_27152049
|
0.018
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr21_-_30631882
|
0.018
|
NM_001077711
|
KRTAP27-1
|
keratin associated protein 27-1
|
|
chr1_-_150463292
|
0.018
|
NM_001009931
|
HRNR
|
hornerin
|
|
chr17_-_39499463
|
0.018
|
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr11_-_5178505
|
0.018
|
NM_001004760
|
OR51V1
|
olfactory receptor, family 51, subfamily V, member 1
|
|
chr4_+_77575276
|
0.017
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr11_+_123945102
|
0.017
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr19_-_55539769
|
0.017
|
|
NAPSB
|
napsin B aspartic peptidase pseudogene
|
|
chr1_-_245942679
|
0.017
|
NM_001005286
|
OR6F1
|
olfactory receptor, family 6, subfamily F, member 1
|
|
chr17_-_36559579
|
0.016
|
NM_033188
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr8_-_17599438
|
0.015
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr1_-_33613729
|
0.015
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr3_+_49522871
|
0.015
|
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr2_-_183095308
|
0.015
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr7_+_142459538
|
0.014
|
NM_001001667
|
OR6V1
|
olfactory receptor, family 6, subfamily V, member 1
|
|
chr4_-_155753249
|
0.013
|
|
FGG
|
fibrinogen gamma chain
|
|
chr11_+_89458765
|
0.013
|
NM_001143975
|
UBTFL1
|
upstream binding transcription factor, RNA polymerase I-like 1
|
|
chr11_-_55684368
|
0.013
|
NM_001004058
|
OR8K5
|
olfactory receptor, family 8, subfamily K, member 5
|
|
chr9_+_81377419
|
0.012
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr1_-_159867781
|
0.012
|
NM_000570
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b)
|
|
chr18_+_19286784
|
0.011
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr14_+_57964462
|
0.011
|
NM_014749
|
KIAA0586
|
KIAA0586
|
|
chr4_+_187424111
|
0.011
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr9_+_96561750
|
0.010
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr1_+_161305558
|
0.010
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr11_-_116168316
|
0.010
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr3_+_110024234
|
0.010
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr17_-_39500512
|
0.009
|
NM_152344
|
LSM12
|
LSM12 homolog (S. cerevisiae)
|
|
chr4_-_170314382
|
0.007
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr6_-_41362370
|
0.007
|
NM_018643
|
TREM1
|
triggering receptor expressed on myeloid cells 1
|
|
chr18_+_59374372
|
0.006
|
NM_080474
|
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12
|
|
chr1_+_184532027
|
0.006
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chr12_-_48902692
|
0.005
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr8_-_17624009
|
0.005
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr21_-_30460841
|
0.005
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr4_-_140280031
|
0.004
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chrX_+_99786044
|
0.003
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr11_-_66201894
|
0.003
|
|
RBM4B
|
RNA binding motif protein 4B
|
|
chr14_+_38653177
|
0.002
|
NM_001009182
NM_001009183
NM_003616
|
SIP1
|
survival of motor neuron protein interacting protein 1
|
|
chr1_+_84382575
|
0.001
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr12_+_10074542
|
0.001
|
NM_207345
|
CLEC9A
|
C-type lectin domain family 9, member A
|
|
chr9_+_140226457
|
0.001
|
NM_001145249
|
FAM157B
|
family with sequence similarity 157, member B
|
|
chr5_-_142795269
|
0.001
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr17_-_26665213
|
0.001
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr8_+_92330691
|
0.000
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|