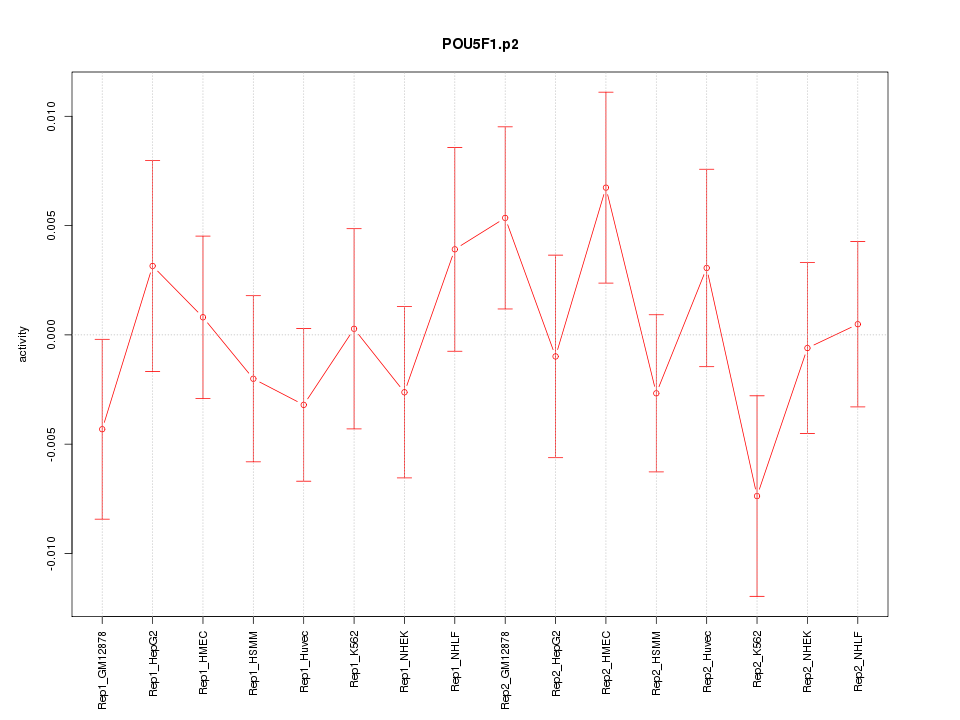
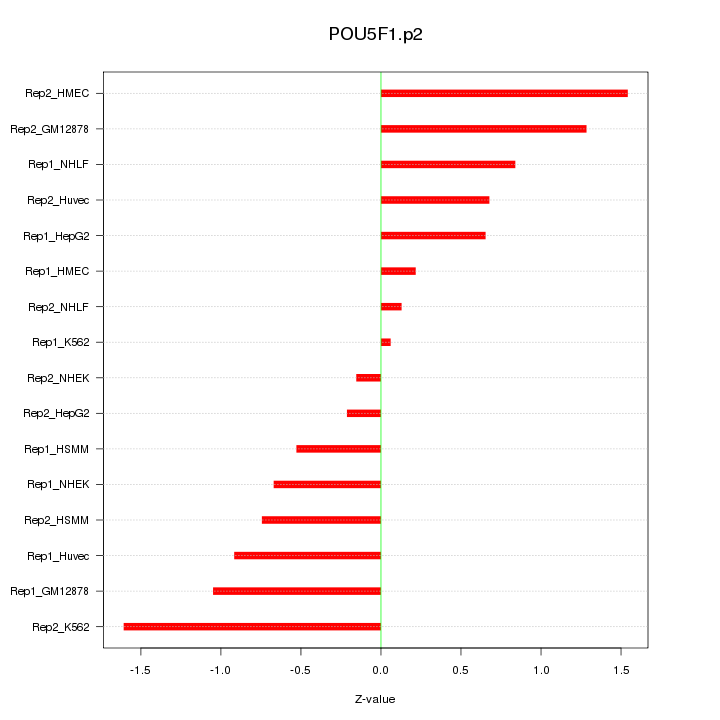
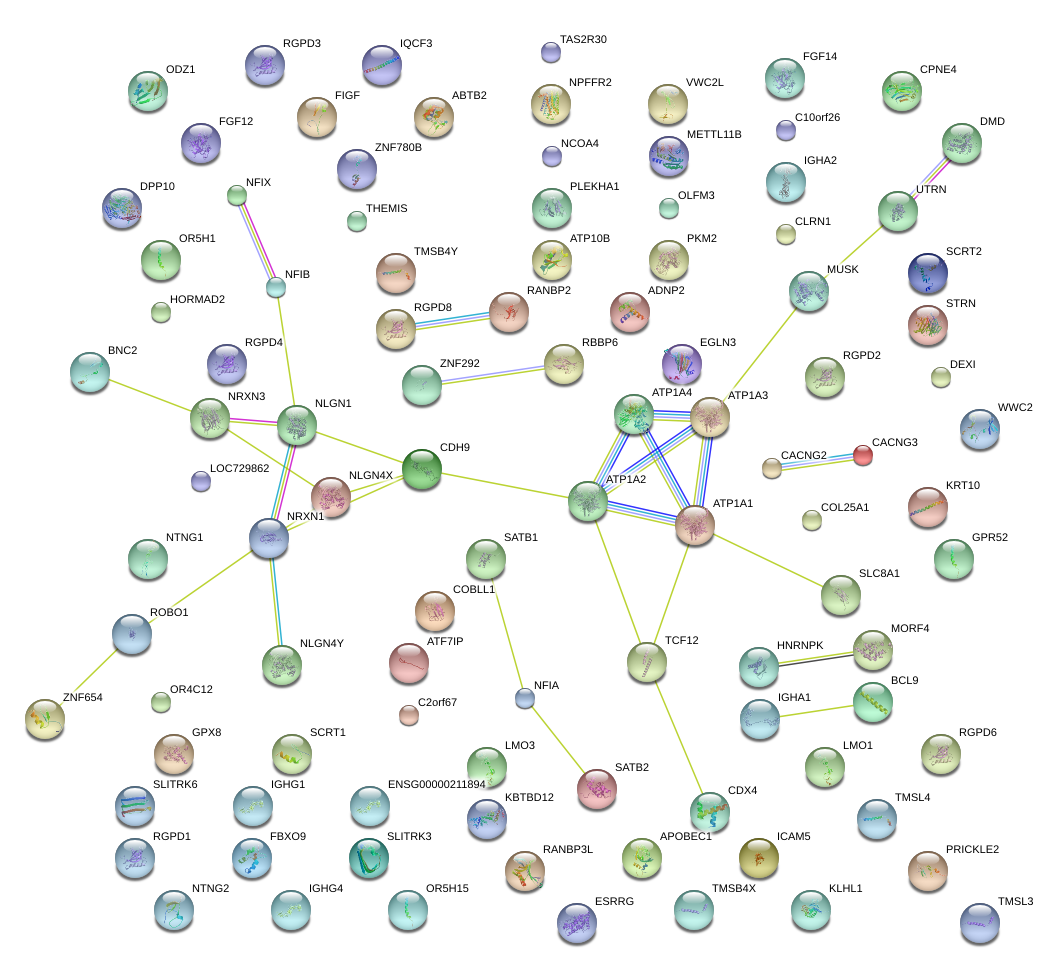
|
chrX_-_33267646
|
1.629
|
NM_000109
|
DMD
|
dystrophin
|
|
chr9_-_14076901
|
1.366
|
|
NFIB
|
nuclear factor I/B
|
|
chr1_-_215329569
|
0.930
|
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr3_+_174598937
|
0.783
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr5_-_27074407
|
0.775
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr2_+_108751033
|
0.674
|
|
RANBP2
|
RAN binding protein 2
|
|
chr4_-_110443247
|
0.665
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr12_-_16649211
|
0.662
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr22_-_35428848
|
0.654
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr2_-_50428395
|
0.651
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr14_+_78815406
|
0.610
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr13_-_101852028
|
0.603
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr2_-_210726539
|
0.600
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr1_+_172683834
|
0.581
|
NM_005684
|
GPR52
|
G protein-coupled receptor 52
|
|
chr3_-_193928038
|
0.575
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr9_-_85773899
|
0.575
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr15_+_26832300
|
0.556
|
|
LOC100289656
|
Dexi homolog (mouse) pseudogene
|
|
chr3_+_129124591
|
0.549
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr12_-_11178109
|
0.547
|
NM_001097643
|
TAS2R30
|
taste receptor, type 2, member 30
|
|
chrX_-_15312390
|
0.546
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chrX_-_123925343
|
0.537
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1(Drosophila)
|
|
chr3_-_152173475
|
0.529
|
NM_001195794
NM_174878
|
CLRN1
|
clarin 1
|
|
chr1_+_168381811
|
0.519
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr3_+_51837608
|
0.514
|
NM_001085479
|
IQCF3
|
IQ motif containing F3
|
|
chr16_+_24475129
|
0.512
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr3_-_79899748
|
0.510
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr12_-_7709734
|
0.494
|
NM_001644
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1
|
|
chr18_+_75968133
|
0.489
|
|
ADNP2
|
ADNP homeobox 2
|
|
chrX_+_72583814
|
0.486
|
NM_005193
|
CDX4
|
caudal type homeobox 4
|
|
chr11_-_34336102
|
0.477
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr2_+_214984705
|
0.456
|
NM_001080500
|
VWC2L
|
von Willebrand factor C domain containing protein 2-like
|
|
chr4_-_174774368
|
0.455
|
NM_006792
|
MORF4
|
mortality factor 4
|
|
chr10_-_37931864
|
0.452
|
NM_001190489
|
MTRNR2L7
|
MT-RNR2-like 7
|
|
chr17_-_36232366
|
0.444
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr6_-_128263868
|
0.443
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr14_-_106184988
|
0.442
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_16717887
|
0.433
|
|
BNC2
|
basonuclin 2
|
|
chr10_+_124135574
|
0.427
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr1_+_145479805
|
0.426
|
NM_004326
|
BCL9
|
B-cell CLL/lymphoma 9
|
|
chr3_-_64186151
|
0.418
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr2_-_165259334
|
0.410
|
|
COBLL1
|
COBL-like 1
|
|
chr14_-_105997732
|
0.409
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_-_51112743
|
0.409
|
|
NRXN1
|
neurexin 1
|
|
chr1_+_116727514
|
0.403
|
NM_001160234
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr10_+_51246290
|
0.400
|
NM_001145262
|
NCOA4
|
nuclear receptor coactivator 4
|
|
chr10_+_104525877
|
0.396
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr22_+_28806452
|
0.391
|
NM_152510
|
HORMAD2
|
HORMA domain containing 2
|
|
chr4_+_184257337
|
0.386
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr1_-_102234959
|
0.385
|
|
OLFM3
|
olfactomedin 3
|
|
chr2_+_114935623
|
0.383
|
NM_001178036
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr5_+_54491702
|
0.382
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr11_-_49960646
|
0.372
|
NM_001005270
|
OR4C12
|
olfactory receptor, family 4, subfamily C, member 12
|
|
chr3_-_18455207
|
0.366
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chrX_-_6156655
|
0.366
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr9_+_112470871
|
0.363
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr3_+_99334230
|
0.355
|
NM_001005338
|
OR5H1
|
olfactory receptor, family 5, subfamily H, member 1
|
|
chr14_-_33489708
|
0.354
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr3_-_133236447
|
0.352
|
NM_130808
|
CPNE4
|
copine IV
|
|
chr13_-_69580459
|
0.351
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|
|
chr5_-_160211623
|
0.349
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr2_+_89715037
|
0.349
|
|
|
|
|
chr5_-_36337756
|
0.348
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr13_+_96797290
|
0.348
|
|
|
|
|
chr12_+_14409875
|
0.348
|
NM_018179
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr2_-_36929520
|
0.347
|
|
STRN
|
striatin, calmodulin binding protein
|
|
chr15_+_55298908
|
0.340
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr4_-_91979113
|
0.340
|
NM_183049
|
TMSL3
|
thymosin-like 3
|
|
chr3_+_88270951
|
0.340
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr13_-_85271329
|
0.338
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr3_-_166397142
|
0.337
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr20_-_604822
|
0.337
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr6_+_53043729
|
0.330
|
NM_012347
|
FBXO9
|
F-box protein 9
|
|
chr15_-_70288071
|
0.330
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr2_-_40593074
|
0.328
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr2_-_200038055
|
0.327
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr4_+_73116384
|
0.325
|
NM_004885
|
NPFFR2
|
neuropeptide FF receptor 2
|
|
chr10_-_21847126
|
0.322
|
|
|
|
|
chr11_-_56067308
|
0.322
|
NM_001005245
|
OR5M11
|
olfactory receptor, family 5, subfamily M, member 11
|
|
chr6_+_88021644
|
0.320
|
|
ZNF292
|
zinc finger protein 292
|
|
chr19_-_45253947
|
0.318
|
NM_001005851
|
ZNF780B
|
zinc finger protein 780B
|
|
chr1_+_107485071
|
0.318
|
|
NTNG1
|
netrin G1
|
|
chr6_+_45404031
|
0.317
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chrX_-_77801480
|
0.317
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr5_-_39238818
|
0.316
|
|
FYB
|
FYN binding protein
|
|
chr4_-_184480572
|
0.316
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr14_-_105948734
|
0.315
|
|
LOC100126583
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG4
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 4 (G4m marker)
immunoglobulin heavy constant mu
|
|
chr5_-_43326922
|
0.314
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr1_+_191044791
|
0.314
|
NM_002923
|
RGS2
|
regulator of G-protein signaling 2, 24kDa
|
|
chr2_+_189547358
|
0.313
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chrX_-_105169370
|
0.311
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr13_-_60887210
|
0.309
|
|
PCDH20
|
protocadherin 20
|
|
chrX_-_24575375
|
0.308
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr11_-_114880275
|
0.307
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr1_+_190811479
|
0.302
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr14_-_106352471
|
0.301
|
|
|
|
|
chr2_+_102456193
|
0.300
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr4_+_124540114
|
0.299
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_-_151296611
|
0.297
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr4_-_163304567
|
0.296
|
NM_001128427
NM_001128428
NM_020116
|
FSTL5
|
follistatin-like 5
|
|
chr14_-_106064887
|
0.296
|
|
LOC100126583
IGHV3-48
IGHA1
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy variable 3-48
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr14_+_61532293
|
0.294
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr2_-_210744158
|
0.293
|
NM_152519
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr7_-_38915306
|
0.286
|
NM_014396
NM_080631
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae)
|
|
chr5_+_140411144
|
0.286
|
NM_013340
|
PCDHB1
|
protocadherin beta 1
|
|
chr4_-_116254381
|
0.284
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr3_+_189413414
|
0.283
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_99370233
|
0.282
|
NM_001005515
|
OR5H15
|
olfactory receptor, family 5, subfamily H, member 15
|
|
chrX_-_106846924
|
0.282
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr20_-_39680405
|
0.282
|
NM_032221
|
CHD6
|
chromodomain helicase DNA binding protein 6
|
|
chrX_-_106846684
|
0.281
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr1_+_162795328
|
0.280
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr11_-_51269023
|
0.279
|
NM_001005272
|
OR4A5
|
olfactory receptor, family 4, subfamily A, member 5
|
|
chr5_-_146238336
|
0.279
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr1_-_151280201
|
0.278
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr5_-_54317102
|
0.278
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr14_-_106193403
|
0.278
|
|
IGKV3-20
|
immunoglobulin kappa variable 3-20
|
|
chr2_-_191580567
|
0.278
|
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa
|
|
chrX_-_43717864
|
0.277
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr18_+_75967902
|
0.273
|
NM_014913
|
ADNP2
|
ADNP homeobox 2
|
|
chr20_+_55569542
|
0.272
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr18_+_57308754
|
0.272
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr12_+_54231249
|
0.271
|
NM_001005494
|
OR6C4
|
olfactory receptor, family 6, subfamily C, member 4
|
|
chr1_+_190871868
|
0.270
|
NM_002927
NM_144766
|
RGS13
|
regulator of G-protein signaling 13
|
|
chrX_+_50670474
|
0.269
|
NM_005448
|
BMP15
|
bone morphogenetic protein 15
|
|
chr14_-_105785639
|
0.267
|
|
|
|
|
chr14_-_105644300
|
0.267
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr14_-_106249999
|
0.267
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr1_-_118529303
|
0.267
|
NM_206996
|
SPAG17
|
sperm associated antigen 17
|
|
chr3_-_58497913
|
0.265
|
NM_003500
|
ACOX2
|
acyl-CoA oxidase 2, branched chain
|
|
chr3_-_78749633
|
0.265
|
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr10_-_108448990
|
0.264
|
|
SORCS1
|
sortilin-related VPS10 domain containing receptor 1
|
|
chrX_-_32340291
|
0.263
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr14_-_106149623
|
0.263
|
|
|
|
|
chr10_+_115302767
|
0.260
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr2_-_215953818
|
0.259
|
|
FN1
|
fibronectin 1
|
|
chr14_-_105862080
|
0.258
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr8_-_23768242
|
0.258
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr1_-_181889070
|
0.255
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr11_+_62813947
|
0.255
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr14_-_19553191
|
0.254
|
NM_001004712
|
OR4K14
|
olfactory receptor, family 4, subfamily K, member 14
|
|
chr4_+_70831387
|
0.252
|
NM_001025104
NM_001890
|
CSN1S1
|
casein alpha s1
|
|
chr14_-_19572756
|
0.251
|
NM_001004714
|
OR4K13
|
olfactory receptor, family 4, subfamily K, member 13
|
|
chr7_+_142590633
|
0.251
|
NM_176881
|
TAS2R39
|
taste receptor, type 2, member 39
|
|
chr3_-_102194938
|
0.251
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr16_+_52081836
|
0.249
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chrX_-_130792297
|
0.248
|
|
LOC286467
|
hypothetical LOC286467
|
|
chr2_-_119322228
|
0.247
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr3_+_99465777
|
0.246
|
NM_001005479
|
OR5H6
|
olfactory receptor, family 5, subfamily H, member 6
|
|
chr12_-_6642513
|
0.246
|
NM_001127582
NM_001127583
NM_001127584
NM_001127585
NM_001127586
NM_016162
|
ING4
|
inhibitor of growth family, member 4
|
|
chr2_+_196229715
|
0.245
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr11_-_107969554
|
0.245
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr10_+_95507555
|
0.244
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr7_+_141342147
|
0.244
|
NM_004668
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase)
|
|
chr1_+_103999824
|
0.243
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr11_-_26700121
|
0.242
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr7_-_31347032
|
0.242
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr2_-_165768798
|
0.241
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr10_-_121592020
|
0.241
|
|
MCMBP
|
minichromosome maintenance complex binding protein
|
|
chr10_+_90414125
|
0.241
|
NM_001198828
NM_001198829
NM_001198830
NM_004190
|
LIPF
|
lipase, gastric
|
|
chr8_+_104900591
|
0.240
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr2_+_189547377
|
0.240
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr1_+_200018380
|
0.239
|
|
NAV1
|
neuron navigator 1
|
|
chrX_+_15435351
|
0.239
|
NM_001721
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr12_+_10015274
|
0.237
|
NM_138337
NM_201623
|
CLEC12A
|
C-type lectin domain family 12, member A
|
|
chr11_-_83706027
|
0.236
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr2_-_51113040
|
0.236
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr4_-_47534815
|
0.235
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr4_+_74520796
|
0.234
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr5_+_52131686
|
0.234
|
|
PELO
|
pelota homolog (Drosophila)
|
|
chr12_-_11066436
|
0.234
|
NM_176888
|
TAS2R19
|
taste receptor, type 2, member 19
|
|
chr11_-_558419
|
0.232
|
|
|
|
|
chr2_+_155263338
|
0.231
|
NM_002239
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3
|
|
chr22_+_31080871
|
0.231
|
NM_006604
|
RFPL3
|
ret finger protein-like 3
|
|
chrX_+_144710557
|
0.230
|
NM_001144006
NM_001144008
NM_001144009
NM_001144010
|
SLITRK2
|
SLIT and NTRK-like family, member 2
|
|
chr14_+_38719462
|
0.230
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr2_-_95846109
|
0.230
|
|
LOC150759
|
hypothetical LOC150759
|
|
chr1_+_145892190
|
0.229
|
NM_001097616
|
GPR89C
|
G protein-coupled receptor 89C
|
|
chr1_+_213245507
|
0.229
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr19_-_58052664
|
0.228
|
NM_001008801
NM_199132
|
ZNF468
|
zinc finger protein 468
|
|
chr21_+_30724442
|
0.228
|
NM_181600
|
KRTAP13-4
|
keratin associated protein 13-4
|
|
chrX_+_16051344
|
0.228
|
NM_005314
|
GRPR
|
gastrin-releasing peptide receptor
|
|
chr6_+_7052798
|
0.227
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr14_-_106282128
|
0.226
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_-_18033970
|
0.224
|
NM_175886
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1
|
|
chr2_-_207738858
|
0.224
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chrX_+_2934653
|
0.222
|
NM_001011719
|
ARSH
|
arylsulfatase family, member H
|
|
chr3_-_24511180
|
0.222
|
NM_001128176
NM_000461
NM_001128177
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr6_-_151754337
|
0.221
|
NM_020861
|
ZBTB2
|
zinc finger and BTB domain containing 2
|
|
chr9_+_33016482
|
0.220
|
|
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr8_-_16903911
|
0.219
|
NM_019851
|
FGF20
|
fibroblast growth factor 20
|
|
chr14_-_105681377
|
0.219
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr5_-_146237826
|
0.218
|
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr18_+_61568825
|
0.218
|
|
CDH7
|
cadherin 7, type 2
|
|
chr17_-_36276987
|
0.217
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr21_-_26175068
|
0.217
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr2_+_211166323
|
0.217
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr14_+_19761708
|
0.217
|
NM_001004480
|
OR11H6
|
olfactory receptor, family 11, subfamily H, member 6
|
|
chr11_+_58980986
|
0.216
|
NM_001004708
|
OR4D6
|
olfactory receptor, family 4, subfamily D, member 6
|
|
chr18_+_61569136
|
0.215
|
NM_004361
|
CDH7
|
cadherin 7, type 2
|
|
chr2_-_215998181
|
0.215
|
|
FN1
|
fibronectin 1
|