|
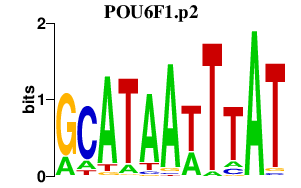
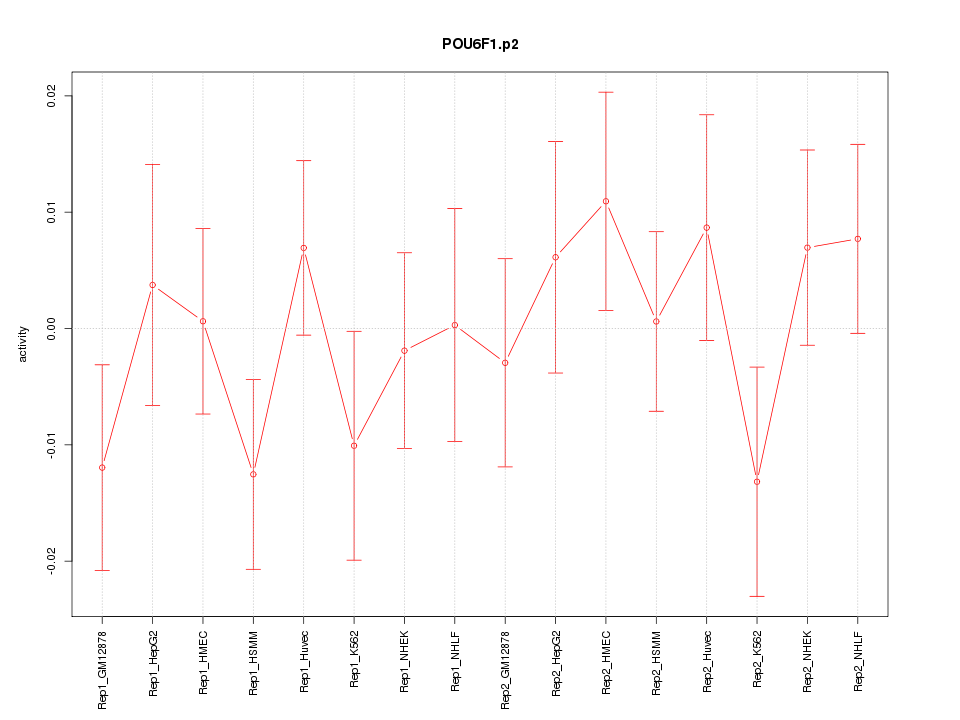
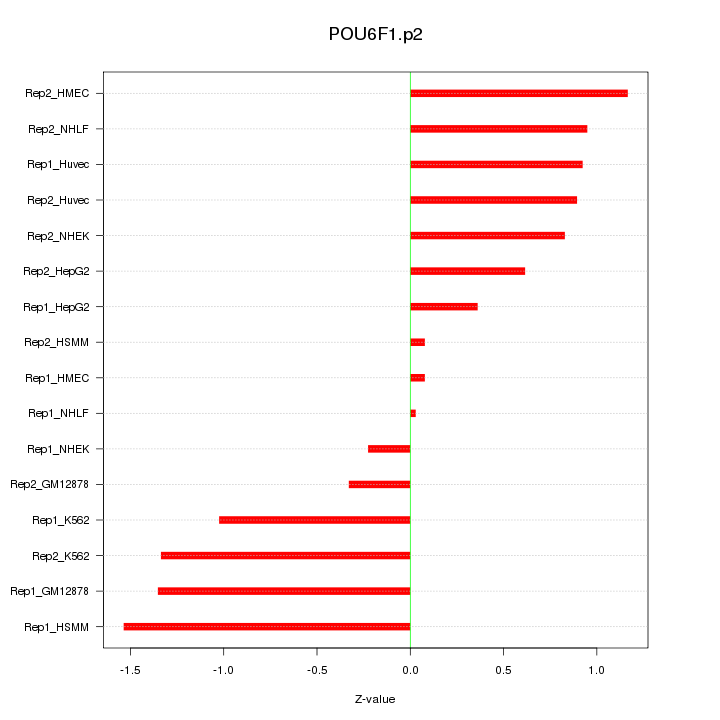
chr8_-_13018123
|
1.895
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr3_-_121117647
|
1.157
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr2_-_72224696
|
0.946
|
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr9_-_106420305
|
0.909
|
NM_001001956
|
OR13C9
|
olfactory receptor, family 13, subfamily C, member 9
|
|
chr4_+_42590038
|
0.909
|
NM_001080476
|
GRXCR1
|
glutaredoxin, cysteine rich 1
|
|
chr14_-_60022487
|
0.868
|
NM_174978
|
C14orf39
|
chromosome 14 open reading frame 39
|
|
chr18_+_59295123
|
0.814
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr3_+_99350859
|
0.760
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr9_+_80134302
|
0.759
|
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr4_+_95595419
|
0.736
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr13_-_25687062
|
0.731
|
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr3_-_191650291
|
0.699
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr21_-_26269298
|
0.696
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr10_+_96433240
|
0.696
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr9_+_19280748
|
0.661
|
NM_017925
|
DENND4C
|
DENN/MADD domain containing 4C
|
|
chr6_-_47362102
|
0.644
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr2_-_225073357
|
0.617
|
|
CUL3
|
cullin 3
|
|
chr5_-_27074407
|
0.614
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr3_-_18455207
|
0.604
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr8_+_17440664
|
0.604
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr5_+_139869819
|
0.580
|
|
|
|
|
chr8_-_67253234
|
0.576
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr14_+_38719462
|
0.558
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr11_-_26550243
|
0.547
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr12_-_11106159
|
0.540
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr7_+_18296569
|
0.535
|
|
HDAC9
|
histone deacetylase 9
|
|
chr2_-_215953818
|
0.533
|
|
FN1
|
fibronectin 1
|
|
chr5_-_160211623
|
0.531
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr20_+_86121
|
0.528
|
NM_139074
|
DEFB127
|
defensin, beta 127
|
|
chr7_+_94135384
|
0.522
|
|
PEG10
|
paternally expressed 10
|
|
chr6_+_127940011
|
0.518
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr3_-_114047479
|
0.516
|
NM_001008784
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr13_+_42253685
|
0.512
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr14_-_56342097
|
0.506
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr8_-_125252884
|
0.504
|
|
C8orf78
|
chromosome 8 open reading frame 78
|
|
chr9_+_21399132
|
0.492
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chr16_-_57309625
|
0.485
|
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chr5_-_95794416
|
0.469
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr2_+_166034402
|
0.467
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr15_+_47502666
|
0.467
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr18_+_45342398
|
0.466
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr3_-_56673113
|
0.463
|
NM_015224
|
C3orf63
|
chromosome 3 open reading frame 63
|
|
chrX_+_28515479
|
0.462
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr5_-_86739683
|
0.462
|
|
CCNH
|
cyclin H
|
|
chr3_+_174598937
|
0.455
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr6_+_53991672
|
0.453
|
NM_138569
|
C6orf142
|
chromosome 6 open reading frame 142
|
|
chr6_-_127822227
|
0.452
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr4_+_146250978
|
0.451
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr18_-_55136673
|
0.450
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr17_-_10313600
|
0.450
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr22_-_38619739
|
0.448
|
NM_152512
|
ENTHD1
|
ENTH domain containing 1
|
|
chr8_-_101384633
|
0.447
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chr14_-_94669547
|
0.443
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr14_+_19473606
|
0.435
|
NM_001004063
|
OR4K1
|
olfactory receptor, family 4, subfamily K, member 1
|
|
chr6_+_54281161
|
0.435
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr1_-_191422346
|
0.418
|
NM_003783
|
B3GALT2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2
|
|
chr6_-_123999640
|
0.418
|
NM_006073
|
TRDN
|
triadin
|
|
chr10_+_4995685
|
0.412
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr14_-_106270497
|
0.410
|
|
IGHG3
|
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr4_-_45820686
|
0.409
|
NM_173536
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1
|
|
chr3_-_121065112
|
0.406
|
|
|
|
|
chr8_-_59575249
|
0.404
|
NM_000780
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1
|
|
chr3_+_129124591
|
0.404
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr3_+_99288706
|
0.404
|
NM_054106
|
OR5AC2
|
olfactory receptor, family 5, subfamily AC, member 2
|
|
chr7_-_38915306
|
0.401
|
NM_014396
NM_080631
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae)
|
|
chr6_-_47361921
|
0.401
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr8_-_93144039
|
0.396
|
|
|
|
|
chr9_-_16717887
|
0.393
|
|
BNC2
|
basonuclin 2
|
|
chr4_-_145281237
|
0.392
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chrX_+_56772391
|
0.369
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr1_+_74473672
|
0.365
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr6_+_144654565
|
0.364
|
NM_007124
|
UTRN
|
utrophin
|
|
chr17_-_10265991
|
0.363
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr12_+_20854904
|
0.358
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr2_-_215949566
|
0.357
|
|
FN1
|
fibronectin 1
|
|
chr4_+_70896236
|
0.355
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chr13_-_102209422
|
0.352
|
NM_001146197
|
C13orf40
|
chromosome 13 open reading frame 40
|
|
chr17_-_36232366
|
0.351
|
NM_000421
|
KRT10
|
keratin 10
|
|
chr1_-_89437098
|
0.350
|
NM_052941
|
GBP4
|
guanylate binding protein 4
|
|
chr14_-_105524207
|
0.349
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr4_-_152368492
|
0.347
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr11_+_55799604
|
0.346
|
NM_001004745
|
OR5T1
|
olfactory receptor, family 5, subfamily T, member 1
|
|
chr7_+_93373755
|
0.342
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr21_-_26345294
|
0.340
|
|
|
|
|
chr3_+_99555387
|
0.339
|
NM_001005517
|
OR5K4
|
olfactory receptor, family 5, subfamily K, member 4
|
|
chr13_+_108046500
|
0.333
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr14_-_100420936
|
0.328
|
NM_001134888
|
RTL1
|
retrotransposon-like 1
|
|
chr4_-_44348414
|
0.324
|
NM_182592
|
YIPF7
|
Yip1 domain family, member 7
|
|
chr4_-_87247829
|
0.322
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr11_-_57927457
|
0.322
|
NM_001005469
|
OR5B3
|
olfactory receptor, family 5, subfamily B, member 3
|
|
chr5_+_140200995
|
0.318
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr6_+_72983183
|
0.316
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_213245507
|
0.312
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr10_+_104493701
|
0.310
|
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr3_-_57301749
|
0.309
|
NM_001142733
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr15_-_100280784
|
0.296
|
NM_001004195
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr4_+_90252990
|
0.292
|
NM_145715
|
TIGD2
|
tigger transposable element derived 2
|
|
chr5_-_145232723
|
0.292
|
NM_001080516
|
GRXCR2
|
glutaredoxin, cysteine rich 2
|
|
chr9_+_5440455
|
0.289
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr2_+_88966170
|
0.288
|
|
|
|
|
chr12_-_44919818
|
0.287
|
|
SLC38A1
|
solute carrier family 38, member 1
|
|
chr12_-_16652202
|
0.282
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr10_+_104493621
|
0.282
|
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr2_-_21081694
|
0.280
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr2_+_166137091
|
0.274
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr12_-_11041740
|
0.272
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr6_-_50045296
|
0.271
|
NM_001037729
|
DEFB113
|
defensin, beta 113
|
|
chr12_-_14024187
|
0.268
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr7_+_119700924
|
0.268
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr3_+_162697289
|
0.267
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr11_-_59390518
|
0.265
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr5_+_161207262
|
0.264
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr11_-_16454493
|
0.262
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr14_-_57030227
|
0.260
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr12_-_11075272
|
0.260
|
NM_176885
|
TAS2R31
|
taste receptor, type 2, member 31
|
|
chr1_+_185064654
|
0.260
|
NM_024420
|
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chrX_-_77801480
|
0.256
|
NM_152694
|
ZCCHC5
|
zinc finger, CCHC domain containing 5
|
|
chr7_+_113853790
|
0.250
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr2_+_210152399
|
0.250
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr11_-_27637771
|
0.246
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_+_147775652
|
0.242
|
|
FBXO38
|
F-box protein 38
|
|
chr13_-_45577144
|
0.233
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr8_+_32698892
|
0.229
|
NM_001159996
|
NRG1
|
neuregulin 1
|
|
chr6_+_72653126
|
0.227
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr10_+_4995453
|
0.226
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr17_-_37794017
|
0.226
|
NM_003150
NM_139276
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor)
|
|
chr3_-_112796855
|
0.225
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr6_+_7524691
|
0.223
|
|
DSP
|
desmoplakin
|
|
chr19_+_61624
|
0.223
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chrX_+_41468351
|
0.222
|
NM_080817
|
GPR82
|
G protein-coupled receptor 82
|
|
chr1_+_103999663
|
0.221
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr3_+_70068440
|
0.221
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr11_+_48284350
|
0.220
|
NM_001004725
|
OR4S1
|
olfactory receptor, family 4, subfamily S, member 1
|
|
chr6_+_163904564
|
0.217
|
|
QKI
|
quaking homolog, KH domain RNA binding (mouse)
|
|
chr15_+_52092392
|
0.216
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr16_+_14201687
|
0.215
|
|
MKL2
|
MKL/myocardin-like 2
|
|
chr2_-_138762909
|
0.213
|
|
|
|
|
chr1_-_214963279
|
0.211
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_-_128199986
|
0.210
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr10_+_4995597
|
0.210
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr17_-_2943039
|
0.208
|
NM_002548
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2
|
|
chr2_+_200878848
|
0.207
|
NM_015535
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like
|
|
chr6_-_29163027
|
0.201
|
NM_001005226
|
OR2B3
|
olfactory receptor, family 2, subfamily B, member 3
|
|
chr17_-_46479127
|
0.201
|
|
SPAG9
|
sperm associated antigen 9
|
|
chrX_+_78087484
|
0.199
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr4_-_145159889
|
0.198
|
NM_002100
|
GYPB
|
glycophorin B (MNS blood group)
|
|
chr8_+_17874128
|
0.198
|
|
PCM1
|
pericentriolar material 1
|
|
chr12_+_98565663
|
0.198
|
NM_153364
|
FAM71C
|
family with sequence similarity 71, member C
|
|
chr1_-_178100204
|
0.197
|
|
|
|
|
chr3_+_35658852
|
0.196
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr7_+_94130673
|
0.194
|
|
PEG10
|
paternally expressed 10
|
|
chr17_-_10362583
|
0.192
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr3_-_110517929
|
0.191
|
NM_138815
|
DPPA2
|
developmental pluripotency associated 2
|
|
chr14_-_105589467
|
0.189
|
|
LOC100126583
IGHA1
IGHG1
IGHG2
IGHM
|
hypothetical LOC100126583
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant mu
|
|
chr16_-_31347151
|
0.189
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr5_+_76284435
|
0.189
|
NM_001882
|
CRHBP
|
corticotropin releasing hormone binding protein
|
|
chr15_+_38082041
|
0.188
|
|
EIF2AK4
|
eukaryotic translation initiation factor 2 alpha kinase 4
|
|
chr4_-_164614335
|
0.187
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr4_+_176076083
|
0.187
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr6_+_64414389
|
0.187
|
NM_015153
|
PHF3
|
PHD finger protein 3
|
|
chr14_-_106289751
|
0.187
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr9_-_124981125
|
0.186
|
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr12_+_16391342
|
0.186
|
NM_145792
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr20_-_218957
|
0.185
|
|
C20orf96
|
chromosome 20 open reading frame 96
|
|
chr17_-_36276987
|
0.184
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr3_-_153470022
|
0.183
|
|
LOC401093
|
hypothetical LOC401093
|
|
chr11_-_55661769
|
0.183
|
NM_001004064
|
OR8J3
|
olfactory receptor, family 8, subfamily J, member 3
|
|
chr1_-_196776697
|
0.180
|
NM_133262
NM_133326
|
ATP6V1G3
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3
|
|
chr6_-_132980901
|
0.179
|
NM_014626
|
TAAR2
|
trace amine associated receptor 2
|
|
chr3_-_115380539
|
0.177
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr1_+_58899
|
0.177
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr2_-_121964373
|
0.176
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr20_+_71230
|
0.176
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr14_-_105523735
|
0.175
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_+_135058402
|
0.175
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr17_-_36294962
|
0.173
|
NM_019010
|
KRT20
|
keratin 20
|
|
chrX_+_46938215
|
0.173
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr11_+_19328846
|
0.169
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr2_-_231697999
|
0.169
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr11_-_57173823
|
0.168
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr15_+_49988882
|
0.168
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr2_+_183290912
|
0.167
|
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr17_-_15914485
|
0.166
|
|
NCOR1
|
nuclear receptor corepressor 1
|
|
chr1_+_93830106
|
0.165
|
|
LOC100129046
|
hypothetical LOC100129046
|
|
chr7_+_91762227
|
0.164
|
|
|
|
|
chr4_+_74825138
|
0.158
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr2_-_27215781
|
0.156
|
NM_178553
|
C2orf53
|
chromosome 2 open reading frame 53
|
|
chr8_-_93177057
|
0.155
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_-_106119742
|
0.154
|
|
IGLJ3
IGHG1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_+_90474280
|
0.153
|
NM_001080518
|
LIPK
|
lipase, family member K
|
|
chr11_-_58736897
|
0.152
|
NM_001039396
|
MPEG1
|
macrophage expressed 1
|
|
chr3_+_100840129
|
0.152
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr14_-_105862080
|
0.152
|
|
IGHV4-31
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr2_+_128009782
|
0.151
|
NM_001080527
|
MYO7B
|
myosin VIIB
|
|
chr8_+_36760999
|
0.151
|
NM_001031836
|
KCNU1
|
potassium channel, subfamily U, member 1
|
|
chr8_-_114518194
|
0.150
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr6_-_138231006
|
0.150
|
|
|
|
|
chr2_-_127817143
|
0.149
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chrX_+_55760774
|
0.149
|
NM_006064
NM_016656
|
RRAGB
|
Ras-related GTP binding B
|
|
chr2_-_225078931
|
0.149
|
|
CUL3
|
cullin 3
|