|
chr22_+_21817512
|
2.515
|
NM_004914
|
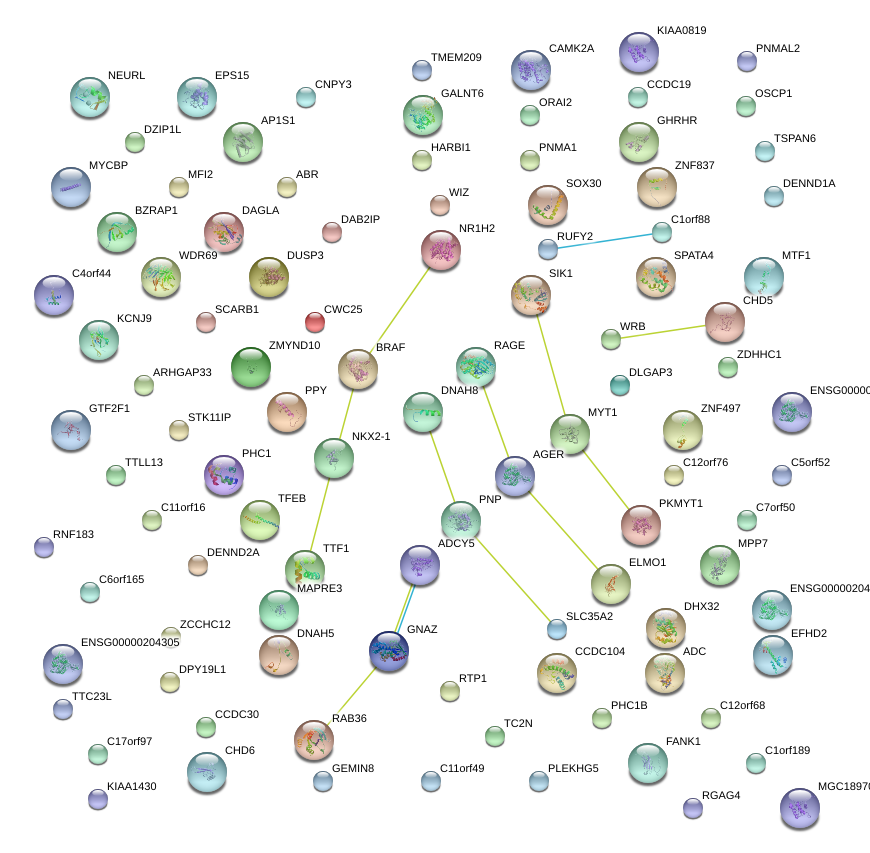
RAB36
|
RAB36, member RAS oncogene family
|
|
chr5_-_149649593
|
2.283
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr5_-_149649380
|
1.986
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr2_+_27046952
|
1.891
|
NM_012326
|
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3
|
|
chr1_+_42701460
|
1.890
|
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr17_-_53761119
|
1.710
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr1_-_158136521
|
1.709
|
NM_012337
|
CCDC19
|
coiled-coil domain containing 19
|
|
chr17_+_53761293
|
1.585
|
|
LOC100506779
|
hypothetical LOC100506779
|
|
chr17_-_959007
|
1.569
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr7_-_139987044
|
1.529
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr1_-_38097811
|
1.269
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr3_-_139315011
|
1.216
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr1_+_33319287
|
1.188
|
NM_052998
|
ADC
|
arginine decarboxylase
|
|
chr3_+_188397967
|
1.091
|
NM_153708
|
RTP1
|
receptor (chemosensory) transporter protein 1
|
|
chr1_-_152445432
|
1.080
|
NM_001010979
|
C1orf189
|
chromosome 1 open reading frame 189
|
|
chr11_+_61204392
|
1.074
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr14_-_73250094
|
1.070
|
NM_006029
|
PNMA1
|
paraneoplastic antigen MA1
|
|
chr22_+_21742364
|
1.051
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr14_+_20007377
|
0.982
|
NM_000270
|
PNP
|
purine nucleoside phosphorylase
|
|
chr7_+_101860958
|
0.963
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr9_-_115101140
|
0.945
|
NM_145051
|
RNF183
|
ring finger protein 183
|
|
chr14_-_101841222
|
0.916
|
NM_014226
|
RAGE
|
renal tumor antigen
|
|
chr7_+_101861007
|
0.913
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr5_-_14064846
|
0.878
|
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr5_+_34875025
|
0.872
|
NM_144725
|
TTC23L
|
tetratricopeptide repeat domain 23-like
|
|
chr19_-_51690989
|
0.860
|
NM_020709
|
PNMAL2
|
PNMA-like 2
|
|
chr17_-_39211706
|
0.859
|
NM_004090
|
DUSP3
|
dual specificity phosphatase 3
|
|
chr22_+_21742727
|
0.840
|
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr1_+_158317967
|
0.824
|
NM_004983
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9
|
|
chr12_+_46863632
|
0.820
|
NM_001013635
|
C12orf68
|
chromosome 12 open reading frame 68
|
|
chr1_+_111690717
|
0.804
|
NM_181643
|
C1orf88
|
chromosome 1 open reading frame 88
|
|
chr1_-_6468108
|
0.802
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr1_-_6480058
|
0.783
|
NM_001042663
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr1_-_36688556
|
0.778
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chrX_-_99778340
|
0.765
|
NM_003270
|
TSPAN6
|
tetraspanin 6
|
|
chr19_+_40958206
|
0.757
|
NM_001172630
NM_052948
|
ARHGAP33
|
Rho GTPase activating protein 33
|
|
chr22_-_16887151
|
0.744
|
NM_001122731
NM_001136004
NM_015241
|
MICAL3
|
microtubule associated monoxygenase, calponin and LIM domain containing 3
|
|
chr1_-_35143570
|
0.740
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr7_-_140270749
|
0.734
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chrX_-_71268475
|
0.733
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr6_+_43005214
|
0.728
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr1_-_6480035
|
0.727
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr9_-_134271985
|
0.723
|
NM_007344
|
TTF1
|
transcription termination factor, RNA polymerase I
|
|
chr5_+_157031138
|
0.719
|
NM_001145132
|
C5orf52
|
chromosome 5 open reading frame 52
|
|
chr21_-_43671355
|
0.705
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr4_-_177353707
|
0.701
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr17_+_260408
|
0.687
|
NM_001013672
|
C17orf97
|
chromosome 17 open reading frame 97
|
|
chr5_-_157031022
|
0.682
|
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr10_+_127575097
|
0.677
|
NM_145235
|
FANK1
|
fibronectin type III and ankyrin repeat domains 1
|
|
chr7_-_129632452
|
0.664
|
NM_032842
|
TMEM209
|
transmembrane protein 209
|
|
chr9_-_125732005
|
0.659
|
NM_020946
NM_024820
|
DENND1A
|
DENN/MADD domain containing 1A
|
|
chrX_-_48653852
|
0.650
|
NM_005660
NM_001032289
|
SLC35A2
|
solute carrier family 35 (UDP-galactose transporter), member A2
|
|
chr19_-_15421761
|
0.646
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr10_-_127574957
|
0.642
|
|
DHX32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32
|
|
chr1_-_6162669
|
0.640
|
NM_015557
|
CHD5
|
chromodomain helicase DNA binding protein 5
|
|
chr1_-_51660328
|
0.640
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr19_-_6344005
|
0.624
|
NM_002096
|
GTF2F1
|
general transcription factor IIF, polypeptide 1, 74kDa
|
|
chr19_+_55571493
|
0.623
|
NM_007121
|
NR1H2
|
nuclear receptor subfamily 1, group H, member 2
|
|
chr12_-_108970658
|
0.620
|
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr2_+_228444570
|
0.619
|
NM_178821
|
WDR69
|
WD repeat domain 69
|
|
chr1_-_39111422
|
0.613
|
NM_012333
|
MYCBP
|
c-myc binding protein
|
|
chr4_+_3220564
|
0.612
|
NM_001042690
|
C4orf44
|
chromosome 4 open reading frame 44
|
|
chr4_-_186362066
|
0.611
|
NM_020827
|
KIAA1430
|
KIAA1430
|
|
chr1_+_15609009
|
0.602
|
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr9_+_123501186
|
0.600
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr10_-_69836912
|
0.592
|
NM_017987
NM_001042417
|
RUFY2
|
RUN and FYVE domain containing 2
|
|
chrX_-_13957879
|
0.584
|
NM_001042479
NM_001042480
NM_017856
|
GEMIN8
|
gem (nuclear organelle) associated protein 8
|
|
chr2_+_220170804
|
0.583
|
NM_052902
|
STK11IP
|
serine/threonine kinase 11 interacting protein
|
|
chr6_+_43005126
|
0.580
|
|
CNPY3
|
canopy 3 homolog (zebrafish)
|
|
chr12_+_8958397
|
0.579
|
NM_004426
|
PHC1
|
polyhomeotic homolog 1 (Drosophila)
|
|
chr11_+_46914804
|
0.578
|
NM_001003676
NM_001003677
NM_001003678
NM_024113
|
C11orf49
|
chromosome 11 open reading frame 49
|
|
chr19_-_63565890
|
0.575
|
NM_198458
|
ZNF497
|
zinc finger protein 497
|
|
chr3_+_184648089
|
0.570
|
|
LOC100505687
|
hypothetical LOC100505687
|
|
chrX_+_117841761
|
0.569
|
NM_173798
|
ZCCHC12
|
zinc finger, CCHC domain containing 12
|
|
chr3_-_50357994
|
0.567
|
NM_015896
|
ZMYND10
|
zinc finger, MYND-type containing 10
|
|
chr10_+_105243724
|
0.566
|
NM_004210
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr7_-_1144347
|
0.565
|
NM_001134395
NM_001134396
NM_032350
|
C7orf50
|
chromosome 7 open reading frame 50
|
|
chr1_+_15608974
|
0.563
|
NM_024329
|
EFHD2
|
EF-hand domain family, member D2
|
|
chr11_-_8911110
|
0.558
|
NM_020643
|
C11orf16
|
chromosome 11 open reading frame 16
|
|
chr15_+_88593765
|
0.553
|
NM_001029964
|
TTLL13
|
tubulin tyrosine ligase-like family, member 13
|
|
chr7_+_100584397
|
0.549
|
NM_001283
|
AP1S1
|
adaptor-related protein complex 1, sigma 1 subunit
|
|
chr6_+_88174408
|
0.548
|
NM_001031743
|
C6orf165
|
chromosome 6 open reading frame 165
|
|
chr2_+_55600220
|
0.544
|
NM_080667
|
CCDC104
|
coiled-coil domain containing 104
|
|
chr7_-_35044177
|
0.542
|
NM_015283
|
DPY19L1
|
dpy-19-like 1 (C. elegans)
|
|
chr7_-_36992221
|
0.538
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr19_-_63584194
|
0.538
|
NM_001129730
NM_138466
|
ZNF837
|
zinc finger protein 837
|
|
chr17_-_34235078
|
0.534
|
NM_017748
|
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chr17_-_34234931
|
0.534
|
|
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chrX_-_48654178
|
0.523
|
NM_001042498
|
SLC35A2
|
solute carrier family 35 (UDP-galactose transporter), member A2
|
|
chr6_-_41811323
|
0.517
|
|
TFEB
|
transcription factor EB
|
|
chr16_-_66007650
|
0.516
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr17_-_55511056
|
0.512
|
NM_022070
|
HEATR6
|
HEAT repeat containing 6
|
|
chr3_-_10027770
|
0.509
|
NM_001008737
|
LOC401052
|
hypothetical LOC401052
|
|
chr3_+_42165740
|
0.508
|
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr14_-_105392671
|
0.506
|
|
|
|
|
chr2_-_64224948
|
0.504
|
NM_020651
|
PELI1
|
pellino homolog 1 (Drosophila)
|
|
chr1_-_203592538
|
0.503
|
NM_018203
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr12_-_4624536
|
0.501
|
NM_006422
|
AKAP3
|
A kinase (PRKA) anchor protein 3
|
|
chr19_-_53706644
|
0.500
|
|
|
|
|
chr12_-_46786107
|
0.495
|
|
SENP1
|
SUMO1/sentrin specific peptidase 1
|
|
chr7_-_157054403
|
0.493
|
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr7_+_156126157
|
0.493
|
NM_030936
NM_001184996
|
RNF32
|
ring finger protein 32
|
|
chr15_-_41572660
|
0.492
|
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr5_-_16795336
|
0.491
|
|
MYO10
|
myosin X
|
|
chr11_+_66841885
|
0.485
|
|
LOC100130987
|
similar to hCG1815675
|
|
chr7_+_128861471
|
0.484
|
NM_001134336
NM_020704
|
FAM40B
|
family with sequence similarity 40, member B
|
|
chrX_+_15666312
|
0.480
|
NM_007220
|
CA5B
|
carbonic anhydrase VB, mitochondrial
|
|
chr6_-_121697305
|
0.480
|
NM_152730
|
C6orf170
|
chromosome 6 open reading frame 170
|
|
chr5_-_1165106
|
0.479
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr1_-_220829704
|
0.477
|
NM_005681
NM_139352
|
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa
|
|
chr21_-_42789369
|
0.475
|
NM_080860
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas)
|
|
chr3_+_47819631
|
0.473
|
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30
|
|
chr21_-_36354659
|
0.471
|
NM_017438
|
SETD4
|
SET domain containing 4
|
|
chr19_-_4408789
|
0.469
|
NM_025241
|
UBXN6
|
UBX domain protein 6
|
|
chr15_-_41572558
|
0.465
|
NM_001141979
NM_001141980
|
TP53BP1
|
tumor protein p53 binding protein 1
|
|
chr21_-_36354428
|
0.463
|
NM_001007259
|
SETD4
|
SET domain containing 4
|
|
chr4_+_123873306
|
0.461
|
NM_001178007
NM_152618
|
BBS12
|
Bardet-Biedl syndrome 12
|
|
chr1_+_37795101
|
0.460
|
NM_003462
|
DNALI1
|
dynein, axonemal, light intermediate chain 1
|
|
chr16_+_28893525
|
0.460
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr10_-_127361583
|
0.459
|
NM_001128202
|
C10orf122
|
chromosome 10 open reading frame 122
|
|
chr5_-_179712920
|
0.455
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr1_-_85497903
|
0.454
|
NM_198077
|
C1orf52
|
chromosome 1 open reading frame 52
|
|
chr9_-_139143980
|
0.454
|
|
|
|
|
chr1_-_199390215
|
0.451
|
NM_016456
|
TMEM9
|
transmembrane protein 9
|
|
chr13_+_30404809
|
0.451
|
NM_152325
|
C13orf26
|
chromosome 13 open reading frame 26
|
|
chr19_+_40325993
|
0.450
|
NM_022006
|
FXYD7
|
FXYD domain containing ion transport regulator 7
|
|
chrX_-_150893752
|
0.448
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chrX_+_118913827
|
0.448
|
NM_001008534
NM_001008535
NM_178813
|
AKAP14
|
A kinase (PRKA) anchor protein 14
|
|
chr21_+_45532377
|
0.444
|
|
LOC642852
|
hypothetical LOC642852
|
|
chr22_-_29082880
|
0.440
|
NM_001005409
NM_005877
|
SF3A1
|
splicing factor 3a, subunit 1, 120kDa
|
|
chr7_+_48041632
|
0.440
|
NM_001100159
|
C7orf57
|
chromosome 7 open reading frame 57
|
|
chr16_-_82736186
|
0.440
|
NM_001146051
NM_031463
|
HSDL1
|
hydroxysteroid dehydrogenase like 1
|
|
chrX_-_49852397
|
0.437
|
NM_003886
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr1_-_24612742
|
0.437
|
NM_001199013
NM_001199014
NM_178122
|
C1orf201
|
chromosome 1 open reading frame 201
|
|
chr2_-_62586979
|
0.436
|
NM_198276
|
TMEM17
|
transmembrane protein 17
|
|
chrX_+_53095230
|
0.431
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr3_+_46717826
|
0.430
|
NM_147196
|
TMIE
|
transmembrane inner ear
|
|
chr11_-_67032675
|
0.429
|
NM_005851
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2
|
|
chr1_+_207926172
|
0.428
|
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr11_+_65469690
|
0.426
|
NM_152762
|
TSGA10IP
|
testis specific, 10 interacting protein
|
|
chr6_-_41811916
|
0.425
|
NM_001167827
|
TFEB
|
transcription factor EB
|
|
chr7_-_122985060
|
0.424
|
NM_005000
|
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa
|
|
chr3_+_47819402
|
0.423
|
NM_014966
NM_138615
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30
|
|
chr4_+_1330957
|
0.421
|
NM_020894
|
KIAA1530
|
KIAA1530
|
|
chr1_-_154837855
|
0.419
|
NM_015590
NM_182679
|
GPATCH4
|
G patch domain containing 4
|
|
chr1_-_31883424
|
0.419
|
NM_012392
|
PEF1
|
penta-EF-hand domain containing 1
|
|
chr16_-_674318
|
0.417
|
|
JMJD8
|
jumonji domain containing 8
|
|
chr1_+_32443822
|
0.417
|
NM_001160042
NM_018134
|
IQCC
|
IQ motif containing C
|
|
chr19_+_4861339
|
0.416
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr19_-_45016587
|
0.410
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr7_-_44195405
|
0.408
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr12_+_124377114
|
0.408
|
NM_052907
|
TMEM132B
|
transmembrane protein 132B
|
|
chr7_+_31693155
|
0.404
|
NM_001145123
NM_006658
|
C7orf16
|
chromosome 7 open reading frame 16
|
|
chr10_-_11693535
|
0.402
|
NM_014688
|
USP6NL
|
USP6 N-terminal like
|
|
chr1_-_21982256
|
0.402
|
NM_001032730
NM_032236
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr16_-_65984836
|
0.402
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr12_-_46785682
|
0.400
|
NM_014554
|
SENP1
|
SUMO1/sentrin specific peptidase 1
|
|
chr11_-_64302777
|
0.400
|
NM_004630
NM_201995
NM_201997
NM_201998
|
SF1
|
splicing factor 1
|
|
chr8_-_37916803
|
0.398
|
NM_152413
|
GOT1L1
|
glutamic-oxaloacetic transaminase 1-like 1
|
|
chr10_+_15179187
|
0.395
|
NM_001097590
NM_006414
NM_183005
|
RPP38
|
ribonuclease P/MRP 38kDa subunit
|
|
chr4_-_123010918
|
0.393
|
NM_018190
NM_176824
|
BBS7
|
Bardet-Biedl syndrome 7
|
|
chr6_-_27548438
|
0.392
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr2_-_130619038
|
0.392
|
NM_207310
|
CCDC74B
|
coiled-coil domain containing 74B
|
|
chr10_+_124020810
|
0.392
|
NM_144587
|
BTBD16
|
BTB (POZ) domain containing 16
|
|
chr17_+_65612589
|
0.391
|
NM_170742
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr7_+_150356435
|
0.386
|
NM_007188
|
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8
|
|
chr3_-_180467483
|
0.386
|
NM_001163677
NM_171828
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3
|
|
chr16_-_56393871
|
0.385
|
|
KIFC3
|
kinesin family member C3
|
|
chr19_-_52045890
|
0.385
|
NM_004069
NM_021575
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit
|
|
chr15_+_53488014
|
0.385
|
NM_001198784
|
FLJ27352
|
hypothetical LOC145788
|
|
chr1_-_202647526
|
0.382
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory (inhibitor) subunit 15B
|
|
chr11_-_56859900
|
0.378
|
NM_003146
|
SSRP1
|
structure specific recognition protein 1
|
|
chr11_-_118716244
|
0.378
|
|
MFRP
|
membrane frizzled-related protein
|
|
chr1_+_109458247
|
0.377
|
|
KIAA1324
|
KIAA1324
|
|
chr19_-_19590438
|
0.376
|
NM_025245
|
PBX4
|
pre-B-cell leukemia homeobox 4
|
|
chrX_-_135941498
|
0.376
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr1_-_199390183
|
0.376
|
|
TMEM9
|
transmembrane protein 9
|
|
chr3_-_16281287
|
0.376
|
NM_001047434
NM_206831
|
DPH3
|
DPH3, KTI11 homolog (S. cerevisiae)
|
|
chr16_-_674357
|
0.375
|
NM_001005920
|
JMJD8
|
jumonji domain containing 8
|
|
chr7_-_156125876
|
0.373
|
|
C7orf13
|
chromosome 7 open reading frame 13
|
|
chr16_+_82736365
|
0.373
|
NM_178452
|
LRRC50
|
leucine rich repeat containing 50
|
|
chr9_+_135314907
|
0.371
|
NM_001135775
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr12_+_120840829
|
0.371
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr1_-_209915443
|
0.368
|
NM_002497
|
NEK2
|
NIMA (never in mitosis gene a)-related kinase 2
|
|
chrX_-_48578878
|
0.368
|
NM_013271
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor
|
|
chr15_+_73803351
|
0.367
|
NM_175881
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1
|
|
chr11_+_8016755
|
0.366
|
NM_003320
|
TUB
|
tubby homolog (mouse)
|
|
chr7_+_30777512
|
0.366
|
NM_032222
|
FAM188B
|
family with sequence similarity 188, member B
|
|
chr1_-_21982192
|
0.366
|
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr1_-_6502655
|
0.365
|
NM_198681
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr1_+_149960636
|
0.364
|
NM_001144956
|
RIIAD1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1
|
|
chr10_+_12211586
|
0.364
|
NM_001142627
NM_001142628
NM_018144
|
SEC61A2
|
Sec61 alpha 2 subunit (S. cerevisiae)
|
|
chr11_+_133444029
|
0.363
|
NM_032801
|
JAM3
|
junctional adhesion molecule 3
|
|
chr3_+_38055699
|
0.360
|
NM_007335
NM_007337
|
DLEC1
|
deleted in lung and esophageal cancer 1
|
|
chr17_-_19206475
|
0.360
|
NM_015681
|
B9D1
|
B9 protein domain 1
|
|
chrX_+_37429927
|
0.360
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr16_+_29971971
|
0.357
|
NM_000034
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr4_-_76774594
|
0.350
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr11_+_45899747
|
0.350
|
NM_152312
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr17_-_40339919
|
0.349
|
|
GFAP
|
glial fibrillary acidic protein
|