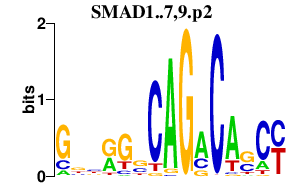
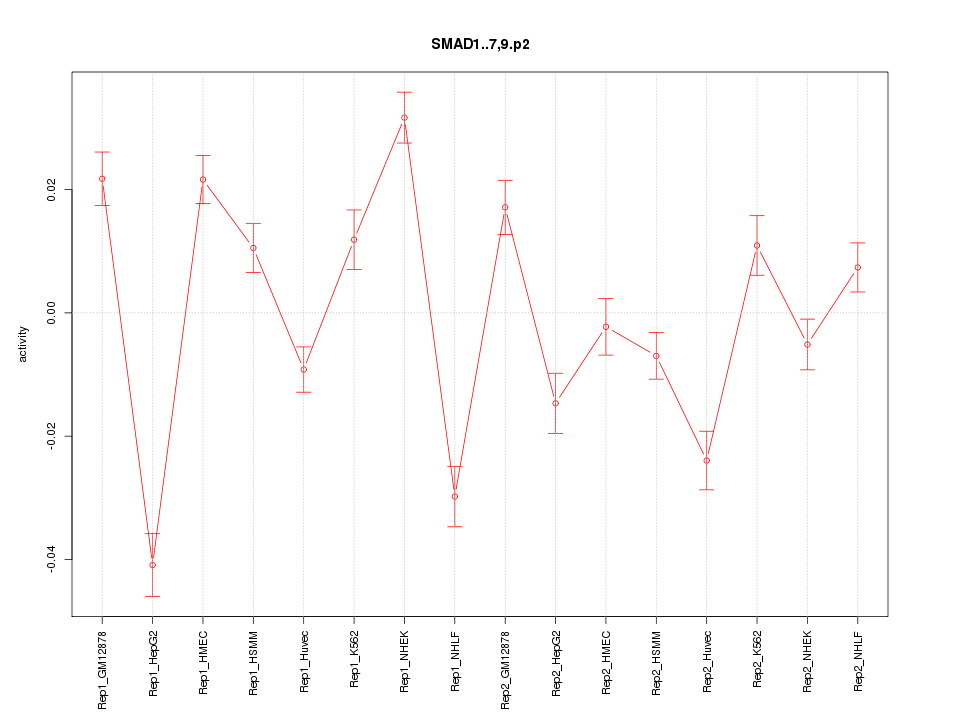
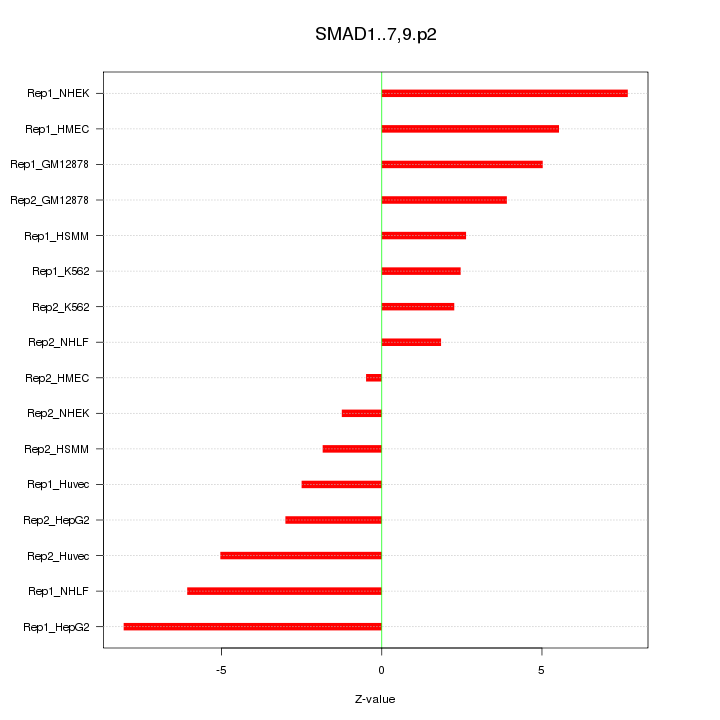
|
chr10_+_135343649
|
7.054
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr10_+_135333667
|
7.004
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191239247
|
6.665
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191226073
|
6.296
|
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
|
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
|
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
|
|
chr10_+_135330357
|
6.228
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191232653
|
5.968
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr4_+_191235959
|
5.914
|
NM_001177376
|
DUX4L4
|
double homeobox 4 like 4
|
|
chrX_+_110226030
|
5.303
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr16_+_56219802
|
5.072
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr11_+_47564805
|
4.961
|
NM_001164379
|
FAM180B
|
family with sequence similarity 180, member B
|
|
chr4_+_191229360
|
4.837
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr9_-_139183675
|
4.515
|
|
|
|
|
chr10_-_102880892
|
4.513
|
NM_001085398
|
TLX1NB
|
TLX1 neighbor
|
|
chrX_+_152682772
|
4.504
|
NM_001163257
NM_005393
|
PLXNB3
|
plexin B3
|
|
chr4_+_191242540
|
4.333
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr19_+_2200112
|
4.241
|
NM_000479
|
AMH
|
anti-Mullerian hormone
|
|
chr17_+_19377732
|
4.221
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr12_+_55896844
|
4.094
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr1_+_15145001
|
4.081
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr8_+_143542378
|
3.934
|
NM_001702
|
BAI1
|
brain-specific angiogenesis inhibitor 1
|
|
chr19_-_1989937
|
3.880
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr22_-_22970850
|
3.843
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr20_+_55399847
|
3.805
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr1_+_8928508
|
3.792
|
NM_001215
|
CA6
|
carbonic anhydrase VI
|
|
chr20_-_61669550
|
3.573
|
NM_033405
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chrX_+_152823722
|
3.487
|
NM_000054
NM_001146151
|
AVPR2
|
arginine vasopressin receptor 2
|
|
chrX_+_135441970
|
3.453
|
NM_016267
|
VGLL1
|
vestigial like 1 (Drosophila)
|
|
chr7_-_150283794
|
3.399
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr8_-_143864933
|
3.394
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr19_-_53808505
|
3.370
|
NM_017708
|
FAM83E
|
family with sequence similarity 83, member E
|
|
chr1_-_6368324
|
3.361
|
NM_181864
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr16_+_87232501
|
3.211
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr21_+_44844924
|
3.129
|
NM_198689
|
KRTAP10-7
|
keratin associated protein 10-7
|
|
chr14_+_99273751
|
3.120
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr11_+_63864263
|
3.116
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr1_+_968522
|
3.110
|
|
AGRN
|
agrin
|
|
chr11_+_303852
|
3.101
|
NM_003641
|
IFITM1
|
interferon induced transmembrane protein 1 (9-27)
|
|
chr8_+_144522389
|
3.085
|
NM_052924
|
RHPN1
|
rhophilin, Rho GTPase binding protein 1
|
|
chr22_+_23221114
|
3.083
|
NM_016327
|
UPB1
|
ureidopropionase, beta
|
|
chr1_-_35795589
|
3.059
|
|
KIAA0319L
|
KIAA0319-like
|
|
chr19_+_60691733
|
2.996
|
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr11_+_113351006
|
2.986
|
NM_000869
NM_213621
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A
|
|
chr17_-_1342700
|
2.975
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chrX_-_102870207
|
2.974
|
NM_001024452
NM_001172285
|
GLRA4
|
glycine receptor, alpha 4
|
|
chr14_+_105012106
|
2.956
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr11_+_74548472
|
2.931
|
NM_001145211
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr19_-_54857003
|
2.922
|
|
IRF3
|
interferon regulatory factor 3
|
|
chr1_-_35795525
|
2.910
|
|
KIAA0319L
|
KIAA0319-like
|
|
chr1_-_35795592
|
2.891
|
NM_024874
|
KIAA0319L
|
KIAA0319-like
|
|
chrX_-_131923092
|
2.866
|
NM_001077188
NM_147175
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr9_-_91284430
|
2.789
|
NM_001142287
NM_006378
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr1_-_1840564
|
2.783
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr7_+_75977680
|
2.755
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chrX_+_79164397
|
2.750
|
NM_016954
|
TBX22
|
T-box 22
|
|
chr3_-_48573556
|
2.749
|
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr8_+_22465368
|
2.708
|
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr9_+_123369219
|
2.655
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr8_+_128817658
|
2.651
|
|
MYC
|
v-myc myelocytomatosis viral oncogene homolog (avian)
|
|
chr10_+_102881050
|
2.628
|
NM_001195517
NM_005521
|
TLX1
|
T-cell leukemia homeobox 1
|
|
chr16_+_2138688
|
2.606
|
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr3_-_48576204
|
2.603
|
NM_033199
|
UCN2
|
urocortin 2
|
|
chr11_-_2136129
|
2.593
|
|
|
|
|
chr9_+_133154896
|
2.592
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr19_+_10392128
|
2.577
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr17_-_24357014
|
2.561
|
NM_001098635
NM_178860
|
SEZ6
|
seizure related 6 homolog (mouse)
|
|
chr7_+_1543644
|
2.542
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr11_-_116199145
|
2.539
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr11_+_1420250
|
2.526
|
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr1_+_9571518
|
2.441
|
NM_001010866
NM_001130924
|
TMEM201
|
transmembrane protein 201
|
|
chr9_-_138387903
|
2.422
|
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chr1_+_178466064
|
2.410
|
NM_033343
|
LHX4
|
LIM homeobox 4
|
|
chr11_-_60475558
|
2.389
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr12_+_48641545
|
2.382
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr17_-_17421050
|
2.367
|
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr21_+_44818033
|
2.359
|
NM_198687
|
KRTAP10-4
|
keratin associated protein 10-4
|
|
chr6_-_32121784
|
2.354
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr11_+_116580892
|
2.351
|
|
TAGLN
|
transgelin
|
|
chr3_+_50259329
|
2.340
|
NM_001166425
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr15_+_39918302
|
2.316
|
NM_001114633
|
PLA2G4B
|
phospholipase A2, group IVB (cytosolic)
|
|
chr22_+_20067662
|
2.311
|
NM_001128633
|
RIMBP3C
|
RIMS binding protein 3C
|
|
chr1_+_199426575
|
2.299
|
NM_001164586
|
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1
|
|
chr3_-_46693902
|
2.297
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr21_+_44856423
|
2.297
|
NM_198695
|
KRTAP10-8
|
keratin associated protein 10-8
|
|
chr7_-_1562272
|
2.292
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr6_+_50789215
|
2.273
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr19_-_46626474
|
2.272
|
NM_198540
|
B3GNT8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8
|
|
chr16_+_2071631
|
2.248
|
|
TSC2
|
tuberous sclerosis 2
|
|
chr9_-_138772506
|
2.245
|
NM_178469
|
LCN8
|
lipocalin 8
|
|
chr19_-_47261761
|
2.242
|
NM_002088
|
GRIK5
|
glutamate receptor, ionotropic, kainate 5
|
|
chr9_-_138387926
|
2.208
|
NM_052813
NM_052814
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chr1_+_100776280
|
2.208
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr16_+_55553262
|
2.207
|
NM_000078
|
CETP
|
cholesteryl ester transfer protein, plasma
|
|
chr17_+_12633504
|
2.195
|
NM_014859
|
ARHGAP44
|
Rho GTPase activating protein 44
|
|
chr14_-_105163301
|
2.186
|
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker)
|
|
chr21_+_43462209
|
2.185
|
NM_000394
|
CRYAA
|
crystallin, alpha A
|
|
chrX_-_135941498
|
2.181
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr14_+_51850754
|
2.179
|
NM_000956
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa
|
|
chr7_-_100331417
|
2.163
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr8_+_142501188
|
2.159
|
NM_007079
NM_032611
|
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3
|
|
chrX_+_151837064
|
2.156
|
NM_001178113
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr17_-_23965305
|
2.139
|
NM_001174103
|
SGK494
|
uncharacterized serine/threonine-protein kinase SgK494
|
|
chr5_+_170779664
|
2.138
|
|
FGF18
|
fibroblast growth factor 18
|
|
chr19_+_17253453
|
2.124
|
NM_152363
|
ANKLE1
|
ankyrin repeat and LEM domain containing 1
|
|
chr3_-_197103848
|
2.123
|
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr9_-_139010805
|
2.110
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr9_+_125813672
|
2.109
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr9_+_139239464
|
2.107
|
NM_199001
|
C9orf169
|
chromosome 9 open reading frame 169
|
|
chr20_-_62052922
|
2.090
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr17_+_71975224
|
2.087
|
NM_001088
|
AANAT
|
aralkylamine N-acetyltransferase
|
|
chr11_+_63627816
|
2.086
|
NM_013280
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1
|
|
chrX_-_41667174
|
2.074
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr1_-_1171964
|
2.065
|
NM_001014980
|
FAM132A
|
family with sequence similarity 132, member A
|
|
chr5_+_170779264
|
2.060
|
NM_003862
|
FGF18
|
fibroblast growth factor 18
|
|
chr3_+_42702014
|
2.047
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr16_-_265868
|
2.042
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr2_+_71016503
|
2.017
|
NM_001692
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1
|
|
chr8_+_128817494
|
2.013
|
NM_002467
|
MYC
|
v-myc myelocytomatosis viral oncogene homolog (avian)
|
|
chr8_+_22493928
|
2.003
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr19_-_17275083
|
2.003
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr21_+_44881700
|
2.001
|
NM_181688
|
KRTAP10-10
|
keratin associated protein 10-10
|
|
chr2_-_232499202
|
1.994
|
NM_024409
|
NPPC
|
natriuretic peptide C
|
|
chrX_+_101853912
|
1.984
|
NM_001004051
NM_001184874
NM_001184875
NM_001184876
NM_138437
|
GPRASP2
|
G protein-coupled receptor associated sorting protein 2
|
|
chr7_-_72761367
|
1.972
|
|
STX1A
|
syntaxin 1A (brain)
|
|
chr3_-_129689394
|
1.969
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr19_+_60691671
|
1.968
|
NM_001144950
NM_001195267
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr11_+_64737886
|
1.961
|
NM_001004326
|
SLC22A20
|
solute carrier family 22, member 20
|
|
chr7_+_27248688
|
1.947
|
NM_001989
|
EVX1
|
even-skipped homeobox 1
|
|
chr7_+_54577512
|
1.941
|
NM_182546
|
VSTM2A
|
V-set and transmembrane domain containing 2A
|
|
chr1_+_20751518
|
1.927
|
NM_207334
|
FAM43B
|
family with sequence similarity 43, member B
|
|
chr15_-_70385709
|
1.927
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr7_+_75947762
|
1.918
|
NM_001102596
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr9_+_125813849
|
1.914
|
|
LHX2
|
LIM homeobox 2
|
|
chr16_-_772926
|
1.904
|
NM_001025190
|
MSLNL
|
mesothelin-like
|
|
chr19_+_3958690
|
1.892
|
NM_015897
|
PIAS4
|
protein inhibitor of activated STAT, 4
|
|
chr9_-_37455250
|
1.882
|
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr19_-_5735775
|
1.882
|
NM_001134316
|
PRR22
|
proline rich 22
|
|
chr6_+_34590624
|
1.860
|
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr19_-_41034734
|
1.854
|
NM_004646
|
NPHS1
|
nephrosis 1, congenital, Finnish type (nephrin)
|
|
chr21_+_44566063
|
1.844
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr6_+_33486750
|
1.843
|
NM_002636
NM_024165
|
PHF1
|
PHD finger protein 1
|
|
chrX_+_149280486
|
1.837
|
|
MAMLD1
|
mastermind-like domain containing 1
|
|
chr9_+_135491305
|
1.833
|
NM_000787
|
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase)
|
|
chr7_+_1063666
|
1.832
|
NM_138445
|
GPR146
|
G protein-coupled receptor 146
|
|
chr22_+_36365629
|
1.831
|
NM_018957
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr3_+_185762233
|
1.831
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr22_+_20068039
|
1.828
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr1_+_203279050
|
1.823
|
|
CNTN2
|
contactin 2 (axonal)
|
|
chr1_+_2397613
|
1.819
|
NM_014638
|
PLCH2
|
phospholipase C, eta 2
|
|
chr1_-_21868380
|
1.812
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr7_+_1546535
|
1.811
|
|
MAFK
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian)
|
|
chr11_+_76455574
|
1.795
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr2_+_233112680
|
1.789
|
NM_005199
|
CHRNG
|
cholinergic receptor, nicotinic, gamma
|
|
chrX_+_100361588
|
1.786
|
NM_001171184
NM_001939
|
DRP2
|
dystrophin related protein 2
|
|
chr11_-_117200618
|
1.783
|
NM_001127489
NM_001680
|
FXYD2
|
FXYD domain containing ion transport regulator 2
|
|
chr4_-_1192749
|
1.765
|
NM_001199021
|
LOC100130872
SPON2
|
hypothetical LOC100130872
spondin 2, extracellular matrix protein
|
|
chr19_+_40474867
|
1.762
|
NM_002361
NM_080600
|
MAG
|
myelin associated glycoprotein
|
|
chr9_+_130056513
|
1.741
|
|
|
|
|
chrX_+_152454773
|
1.739
|
NM_001001344
NM_021949
|
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3
|
|
chr5_+_133479196
|
1.715
|
NM_001134851
NM_201632
NM_201634
NM_213648
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr19_+_40299058
|
1.707
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr18_+_894943
|
1.698
|
NM_001099733
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary)
|
|
chr8_-_145085616
|
1.694
|
NM_201384
|
PLEC
|
plectin
|
|
chr3_+_52325374
|
1.685
|
NM_015512
|
DNAH1
|
dynein, axonemal, heavy chain 1
|
|
chr7_+_129807467
|
1.682
|
NM_001868
|
CPA1
|
carboxypeptidase A1 (pancreatic)
|
|
chr9_+_91409746
|
1.679
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr8_-_124289107
|
1.657
|
|
|
|
|
chr19_-_40711034
|
1.656
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr11_-_2138984
|
1.655
|
NM_000207
NM_001185097
NM_001185098
NM_001042376
|
INS
INS-IGF2
|
insulin
INS-IGF2 readthrough transcript
|
|
chr22_+_29848923
|
1.653
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr10_+_112247614
|
1.649
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chrX_-_62487693
|
1.649
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chr9_-_37455384
|
1.641
|
NM_014872
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr1_+_15935395
|
1.614
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr1_-_12599934
|
1.607
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr9_+_123083540
|
1.600
|
|
GSN
|
gelsolin
|
|
chrX_+_153279764
|
1.597
|
NM_006013
|
RPL10
|
ribosomal protein L10
|
|
chr11_+_65754476
|
1.595
|
|
PACS1
|
phosphofurin acidic cluster sorting protein 1
|
|
chr3_+_127725851
|
1.593
|
NM_152889
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr19_-_40684638
|
1.592
|
NM_001035516
|
DMKN
|
dermokine
|
|
chrX_-_74062005
|
1.590
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr15_-_50869379
|
1.588
|
NM_004498
|
ONECUT1
|
one cut homeobox 1
|
|
chr11_+_60727559
|
1.584
|
NM_001079807
|
PGA3
PGA4
|
pepsinogen 3, group I (pepsinogen A)
pepsinogen 4, group I (pepsinogen A)
|
|
chr8_+_145692916
|
1.577
|
NM_032902
|
PPP1R16A
|
protein phosphatase 1, regulatory (inhibitor) subunit 16A
|
|
chr1_-_10779291
|
1.575
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|
|
chr19_+_45998020
|
1.573
|
NM_053046
|
EGLN2
|
egl nine homolog 2 (C. elegans)
|
|
chr10_-_71002998
|
1.556
|
NM_020999
|
NEUROG3
|
neurogenin 3
|
|
chr17_-_53761119
|
1.554
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chrX_-_19598912
|
1.552
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chrX_-_153698930
|
1.550
|
|
HMGN1P37
|
high-mobility group nucleosome binding domain 1 pseudogene 37
|
|
chr10_-_70846573
|
1.550
|
NM_001057
|
TACR2
|
tachykinin receptor 2
|
|
chr6_-_2580296
|
1.541
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr12_-_121224676
|
1.541
|
NM_001014336
|
IL31
|
interleukin 31
|
|
chr21_-_41479035
|
1.539
|
NM_182832
|
PLAC4
|
placenta-specific 4
|
|
chr1_-_1139358
|
1.533
|
NM_003327
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4
|
|
chr22_+_21318779
|
1.533
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr3_+_11009419
|
1.520
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr12_+_109956210
|
1.518
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr9_+_125158268
|
1.517
|
NM_173689
|
CRB2
|
crumbs homolog 2 (Drosophila)
|
|
chr1_-_43523825
|
1.517
|
NM_182517
NM_001164829
|
C1orf210
|
chromosome 1 open reading frame 210
|
|
chr16_+_69060325
|
1.505
|
|
FUK
|
fucokinase
|