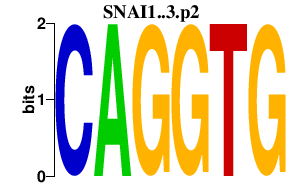
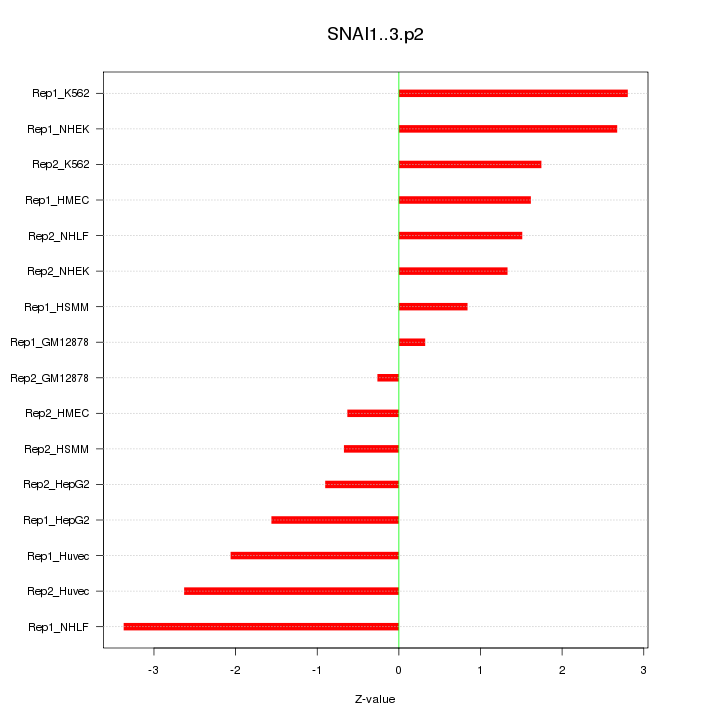
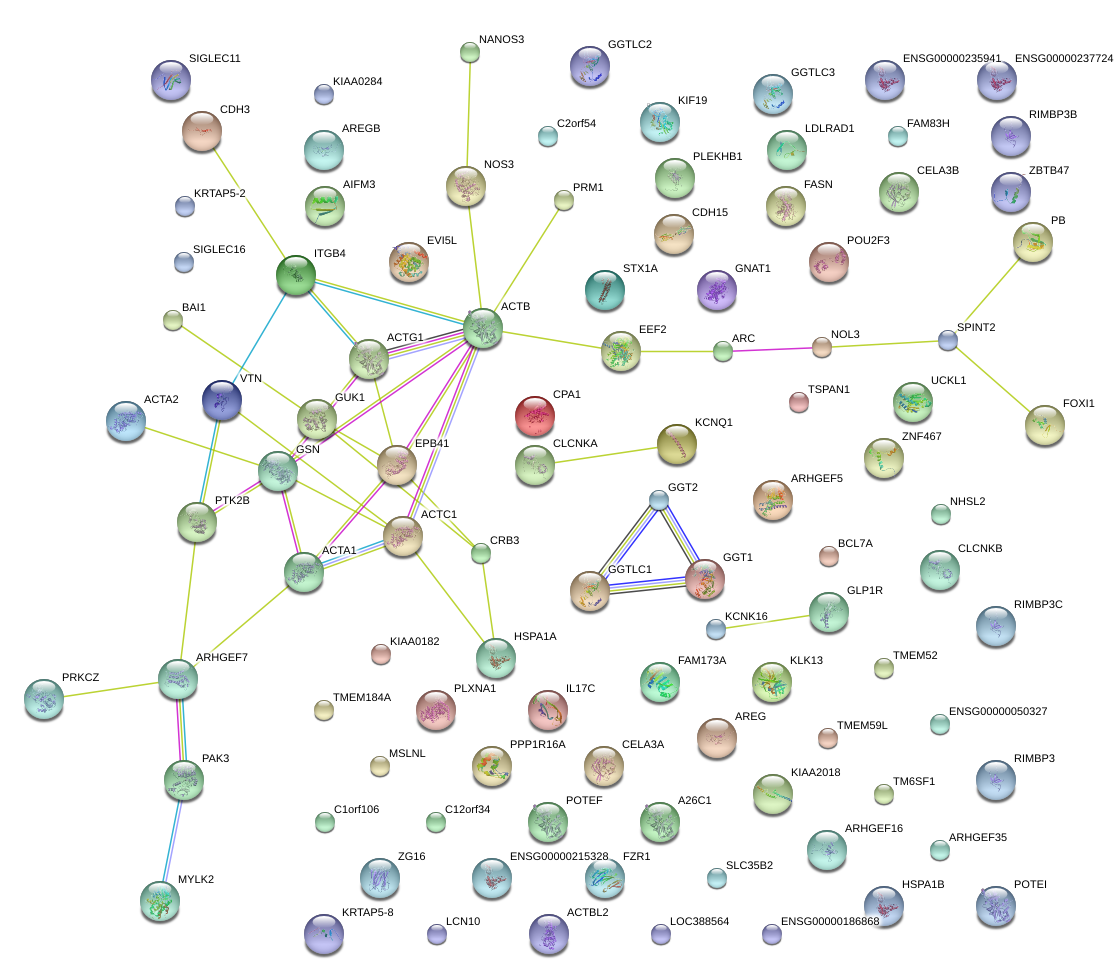
|
chr2_-_241484245
|
3.139
|
NM_001085437
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr7_-_72761367
|
2.749
|
|
STX1A
|
syntaxin 1A (brain)
|
|
chr1_+_3360880
|
2.449
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr8_+_143542378
|
2.437
|
NM_001702
|
BAI1
|
brain-specific angiogenesis inhibitor 1
|
|
chr19_+_43447078
|
2.367
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr19_+_6415259
|
2.287
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr19_-_60587414
|
2.224
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr19_+_43446937
|
2.223
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_-_1840564
|
2.132
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr19_+_43447262
|
2.120
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr6_+_31891293
|
2.054
|
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr6_+_31891509
|
2.031
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr6_+_31903374
|
1.834
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr17_+_69875140
|
1.807
|
NM_181790
|
GPR142
|
G protein-coupled receptor 142
|
|
chr22_+_20067662
|
1.805
|
NM_001128633
|
RIMBP3C
|
RIMS binding protein 3C
|
|
chr1_+_199130571
|
1.793
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr22_+_21027538
|
1.744
|
|
|
|
|
chr16_+_87232501
|
1.719
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr17_-_77635703
|
1.716
|
|
FASN
|
fatty acid synthase
|
|
chr1_+_1062137
|
1.715
|
|
LOC254099
|
hypothetical protein LOC254099
|
|
chr22_+_21318779
|
1.703
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr11_+_119616112
|
1.702
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr14_+_104402626
|
1.684
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr1_+_226399026
|
1.679
|
NM_001159391
|
GUK1
|
guanylate kinase 1
|
|
chr6_-_44333002
|
1.671
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr19_-_3928516
|
1.643
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr8_+_145692916
|
1.634
|
NM_032902
|
PPP1R16A
|
protein phosphatase 1, regulatory (inhibitor) subunit 16A
|
|
chr19_-_3931499
|
1.630
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr17_+_71235324
|
1.619
|
|
ITGB4
|
integrin, beta 4
|
|
chr12_+_120944174
|
1.606
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr5_+_169465478
|
1.591
|
NM_012188
NM_144769
|
FOXI1
|
forkhead box I1
|
|
chr17_-_77092569
|
1.585
|
|
ACTG1
|
actin, gamma 1
|
|
chr1_+_1971762
|
1.576
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr20_-_62052922
|
1.559
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr7_-_149101227
|
1.557
|
NM_207336
|
ZNF467
|
zinc finger protein 467
|
|
chr16_+_84204424
|
1.554
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr20_+_29870836
|
1.530
|
NM_033118
|
MYLK2
|
myosin light chain kinase 2
|
|
chr19_+_55164668
|
1.521
|
|
SIGLEC16
|
sialic acid binding Ig-like lectin 16 (gene/pseudogene)
|
|
chr1_+_29086156
|
1.501
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr1_+_29086206
|
1.501
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr12_+_108690199
|
1.486
|
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr8_-_143692814
|
1.485
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr8_+_27238979
|
1.446
|
NM_173175
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr3_+_50204026
|
1.417
|
NM_000172
NM_144499
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1
|
|
chr8_-_144882679
|
1.411
|
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr3_+_128190070
|
1.405
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr1_+_46418786
|
1.402
|
|
TSPAN1
|
tetraspanin 1
|
|
chr1_+_16220952
|
1.391
|
NM_001042704
NM_004070
|
CLCNKA
CLCNKB
|
chloride channel Ka
chloride channel Kb
|
|
chr4_+_75529716
|
1.390
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr19_+_18584666
|
1.387
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr16_+_711139
|
1.379
|
NM_023933
|
FAM173A
|
family with sequence similarity 173, member A
|
|
chr1_-_54256390
|
1.360
|
NM_001010978
|
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1
|
|
chr6_-_39398293
|
1.330
|
NM_001135105
NM_001135106
NM_001135107
NM_032115
|
KCNK16
|
potassium channel, subfamily K, member 16
|
|
chr16_+_29697090
|
1.321
|
|
ZG16
|
zymogen granule protein 16 homolog (rat)
|
|
chr22_+_21464980
|
1.321
|
|
|
|
|
chr16_+_67236730
|
1.311
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr11_+_73036241
|
1.296
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr9_-_138757175
|
1.292
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr7_-_1562272
|
1.269
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr7_+_150321771
|
1.252
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chrX_+_110226030
|
1.250
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr1_+_22176004
|
1.242
|
NM_007352
|
CELA3A
CELA3B
|
chymotrypsin-like elastase family, member 3A
chymotrypsin-like elastase family, member 3B
|
|
chr19_+_7834124
|
1.241
|
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr22_+_19660361
|
1.237
|
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3
|
|
chr17_-_23716391
|
1.218
|
NM_001080837
|
SEBOX
|
SEBOX homeobox
|
|
chr7_+_129807467
|
1.199
|
NM_001868
|
CPA1
|
carboxypeptidase A1 (pancreatic)
|
|
chr6_+_39124534
|
1.189
|
NM_002062
|
GLP1R
|
glucagon-like peptide 1 receptor
|
|
chr16_-_772926
|
1.182
|
NM_001025190
|
MSLNL
|
mesothelin-like
|
|
chr19_+_3457264
|
1.179
|
NM_016263
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr17_-_77634455
|
1.176
|
|
FASN
|
fatty acid synthase
|
|
chr11_+_2422747
|
1.172
|
NM_000218
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1
|
|
chr16_-_11282595
|
1.161
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr7_+_143683365
|
1.158
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr4_+_75699652
|
1.155
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr3_+_42675841
|
1.147
|
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr19_-_3930828
|
1.145
|
|
EEF2
|
eukaryotic translation elongation factor 2
|
|
chr15_+_81567327
|
1.140
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr11_+_70926718
|
1.138
|
NM_021046
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chrX_+_71047344
|
1.134
|
NM_001013627
|
NHSL2
|
NHS-like 2
|
|
chr10_+_71060008
|
1.121
|
NM_145306
|
C10orf35
|
chromosome 10 open reading frame 35
|
|
chr17_-_77469319
|
1.102
|
|
SIRT7
|
sirtuin 7
|
|
chr5_+_1254709
|
1.099
|
NM_001003841
|
SLC6A19
|
solute carrier family 6 (neutral amino acid transporter), member 19
|
|
chr6_+_43507466
|
1.092
|
NM_033450
|
ABCC10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10
|
|
chr7_-_72822471
|
1.090
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr17_-_77469316
|
1.087
|
|
SIRT7
|
sirtuin 7
|
|
chrX_-_69186506
|
1.074
|
NM_001002254
|
AWAT2
|
acyl-CoA wax alcohol acyltransferase 2
|
|
chr17_-_77469327
|
1.073
|
NM_016538
|
SIRT7
|
sirtuin 7
|
|
chr5_-_525916
|
1.066
|
|
LOC25845
|
hypothetical LOC25845
|
|
chr1_+_24755152
|
1.065
|
NM_001010980
|
C1orf130
|
chromosome 1 open reading frame 130
|
|
chr13_+_112681626
|
1.064
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr22_-_17891777
|
1.054
|
|
CLDN5
|
claudin 5
|
|
chr1_+_2149993
|
1.054
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr8_-_145609737
|
1.053
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr9_+_139153409
|
1.052
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr17_+_24095135
|
1.049
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr16_-_790699
|
1.046
|
NM_016541
|
GNG13
|
guanine nucleotide binding protein (G protein), gamma 13
|
|
chr22_+_21852743
|
1.041
|
|
BCR
|
breakpoint cluster region
|
|
chr1_-_11841476
|
1.040
|
NM_002521
|
NPPB
|
natriuretic peptide B
|
|
chr1_-_204179169
|
1.034
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chrX_-_48654178
|
1.033
|
NM_001042498
|
SLC35A2
|
solute carrier family 35 (UDP-galactose transporter), member A2
|
|
chr1_-_17318452
|
1.030
|
NM_007365
|
PADI2
|
peptidyl arginine deiminase, type II
|
|
chr11_-_2127368
|
1.022
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr22_-_20235749
|
1.018
|
NM_001128633
|
RIMBP3C
|
RIMS binding protein 3C
|
|
chr22_+_18081968
|
1.017
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr16_+_4325142
|
1.012
|
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr11_+_61204392
|
1.010
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr10_+_46413551
|
1.009
|
NM_014696
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2
|
|
chr17_+_26839143
|
1.006
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr17_+_26060617
|
0.994
|
|
|
|
|
chr22_-_48793181
|
0.992
|
NM_001001694
|
IL17REL
|
interleukin 17 receptor E-like
|
|
chrX_+_70359780
|
0.992
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr7_+_75947762
|
0.992
|
NM_001102596
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr19_-_59438411
|
0.989
|
NM_024318
|
LILRB3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr1_-_3437834
|
0.989
|
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr11_+_64631919
|
0.985
|
|
C11orf2
|
chromosome 11 open reading frame 2
|
|
chr1_-_43509187
|
0.985
|
|
EBNA1BP2
|
EBNA1 binding protein 2
|
|
chr6_+_33697023
|
0.977
|
NM_002224
|
ITPR3
|
inositol 1,4,5-triphosphate receptor, type 3
|
|
chr17_-_77129802
|
0.972
|
NM_025161
|
C17orf70
|
chromosome 17 open reading frame 70
|
|
chr5_-_1165106
|
0.971
|
NM_006598
|
SLC12A7
|
solute carrier family 12 (potassium/chloride transporters), member 7
|
|
chr15_-_73818013
|
0.969
|
|
|
|
|
chr15_-_75694674
|
0.969
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr6_+_43319199
|
0.965
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr1_-_1139358
|
0.964
|
NM_003327
|
TNFRSF4
|
tumor necrosis factor receptor superfamily, member 4
|
|
chr22_+_21340757
|
0.958
|
|
|
|
|
chr8_+_27238964
|
0.956
|
NM_173176
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr9_-_130980302
|
0.952
|
|
IER5L
|
immediate early response 5-like
|
|
chr15_-_38332401
|
0.948
|
NM_001039905
|
C15orf56
|
chromosome 15 open reading frame 56
|
|
chr9_+_132961683
|
0.946
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr22_+_21384862
|
0.943
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr19_+_3490154
|
0.940
|
NM_001135580
|
C19orf71
|
chromosome 19 open reading frame 71
|
|
chr16_+_67236699
|
0.940
|
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr17_-_43390108
|
0.934
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr22_+_19248733
|
0.931
|
|
MED15
|
mediator complex subunit 15
|
|
chr11_-_6600385
|
0.927
|
|
DCHS1
|
dachsous 1 (Drosophila)
|
|
chr1_-_55039458
|
0.927
|
NM_001114108
NM_017904
|
TTC22
|
tetratricopeptide repeat domain 22
|
|
chr12_+_111980044
|
0.927
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr7_-_130891861
|
0.919
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr11_+_56984702
|
0.917
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr3_+_42679602
|
0.916
|
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr9_-_139060416
|
0.915
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr2_+_238712101
|
0.915
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr1_-_3517918
|
0.912
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr8_+_102574012
|
0.907
|
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr20_-_60375695
|
0.906
|
NM_005560
|
LAMA5
|
laminin, alpha 5
|
|
chr19_+_45799060
|
0.906
|
NM_001042545
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chrX_-_48661226
|
0.903
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr21_+_44818033
|
0.898
|
NM_198687
|
KRTAP10-4
|
keratin associated protein 10-4
|
|
chr1_+_1104939
|
0.896
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr22_+_21006814
|
0.896
|
|
|
|
|
chr19_-_51829778
|
0.896
|
NM_033258
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8
|
|
chr3_+_129682434
|
0.895
|
|
|
|
|
chr9_-_139060285
|
0.885
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr9_-_130980359
|
0.884
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr11_+_183079
|
0.883
|
NM_145651
|
SCGB1C1
|
secretoglobin, family 1C, member 1
|
|
chr16_+_3002457
|
0.883
|
NM_020982
|
CLDN9
|
claudin 9
|
|
chr22_-_20235372
|
0.883
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr11_+_60448487
|
0.883
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr9_+_132961723
|
0.882
|
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr19_-_2378530
|
0.877
|
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr19_+_15765760
|
0.876
|
NM_001004466
|
OR10H5
|
olfactory receptor, family 10, subfamily H, member 5
|
|
chr9_+_135389698
|
0.876
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr19_-_7896974
|
0.874
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr17_+_73676219
|
0.871
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr22_-_17545872
|
0.869
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr11_+_61279436
|
0.869
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr10_-_98470164
|
0.866
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr19_-_54212158
|
0.859
|
NM_000894
|
LHB
|
luteinizing hormone beta polypeptide
|
|
chr7_-_131911808
|
0.857
|
NM_001105543
NM_020911
|
PLXNA4
|
plexin A4
|
|
chr19_+_51424848
|
0.856
|
NM_198541
|
IGFL1
|
IGF-like family member 1
|
|
chr1_+_226352179
|
0.854
|
|
ARF1
|
ADP-ribosylation factor 1
|
|
chr15_+_72687765
|
0.852
|
NM_003992
|
CLK3
|
CDC-like kinase 3
|
|
chr1_+_154478574
|
0.849
|
NM_199173
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein
|
|
chr1_+_18680010
|
0.845
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr7_+_5598979
|
0.843
|
NM_003088
|
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus)
|
|
chr17_+_18221803
|
0.833
|
NM_001145127
|
EVPLL
|
envoplakin-like
|
|
chr17_-_77469280
|
0.832
|
|
SIRT7
|
sirtuin 7
|
|
chr11_+_1607580
|
0.831
|
NM_001001480
|
KRTAP5-5
|
keratin associated protein 5-5
|
|
chr17_+_4434582
|
0.830
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr14_+_104117100
|
0.825
|
NM_001008404
|
C14orf180
|
chromosome 14 open reading frame 180
|
|
chr1_-_2451543
|
0.823
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr9_+_123544884
|
0.820
|
NM_138709
|
DAB2IP
|
DAB2 interacting protein
|
|
chr1_+_41057186
|
0.819
|
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr21_+_44530148
|
0.817
|
NM_000383
|
AIRE
|
autoimmune regulator
|
|
chr8_-_41641881
|
0.814
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr18_+_19706979
|
0.814
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr17_+_7225088
|
0.814
|
NM_003985
|
TNK1
|
tyrosine kinase, non-receptor, 1
|
|
chr1_-_19058741
|
0.812
|
NM_152232
|
TAS1R2
|
taste receptor, type 1, member 2
|
|
chr16_-_87460514
|
0.812
|
NM_001080487
|
PABPN1L
|
poly(A) binding protein, nuclear 1-like (cytoplasmic)
|
|
chr1_-_2486313
|
0.811
|
NM_003820
|
TNFRSF14
|
tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator)
|
|
chr22_+_20350272
|
0.808
|
NM_014337
NM_148175
NM_148176
|
PPIL2
|
peptidylprolyl isomerase (cyclophilin)-like 2
|
|
chr10_+_43187095
|
0.807
|
NM_001184963
NM_173160
|
FXYD4
|
FXYD domain containing ion transport regulator 4
|
|
chr1_+_31855873
|
0.801
|
NM_001525
|
HCRTR1
|
hypocretin (orexin) receptor 1
|
|
chr22_+_17847467
|
0.800
|
|
CDC45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr19_+_1236766
|
0.800
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr19_+_41039649
|
0.799
|
NM_032123
NM_199179
NM_199180
|
KIRREL2
|
kin of IRRE like 2 (Drosophila)
|
|
chr22_+_18124225
|
0.798
|
NM_005992
NM_080646
NM_080647
|
TBX1
|
T-box 1
|
|
chr1_+_95355415
|
0.798
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr9_-_139060377
|
0.788
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr8_+_22493928
|
0.787
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr22_-_20314366
|
0.786
|
|
YDJC
|
YdjC homolog (bacterial)
|