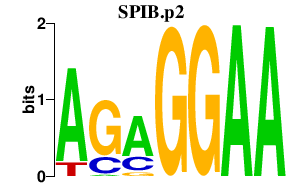
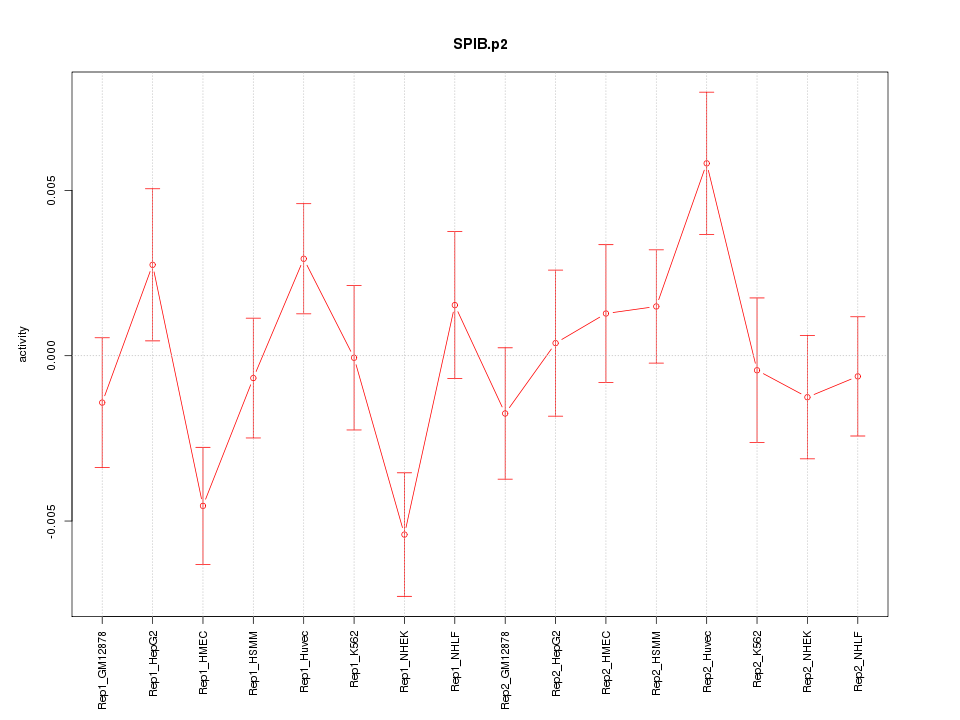
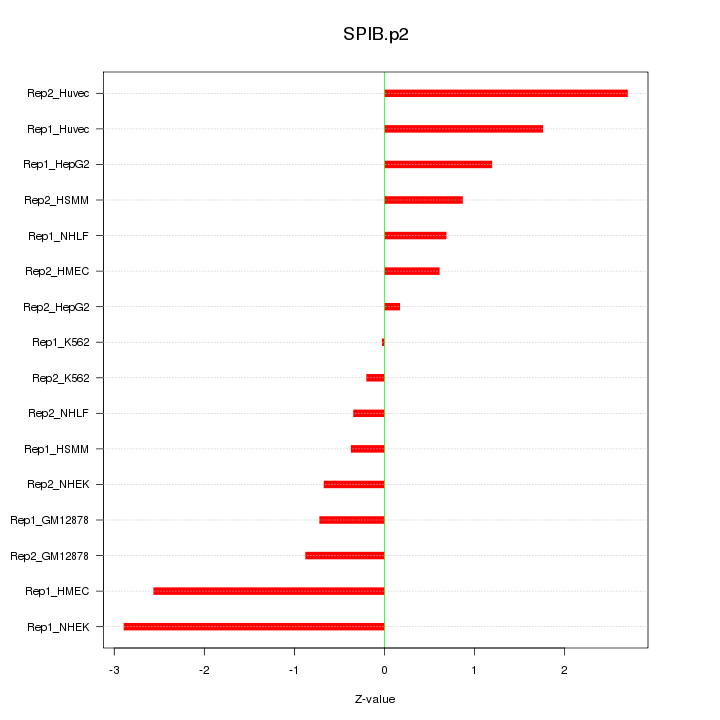
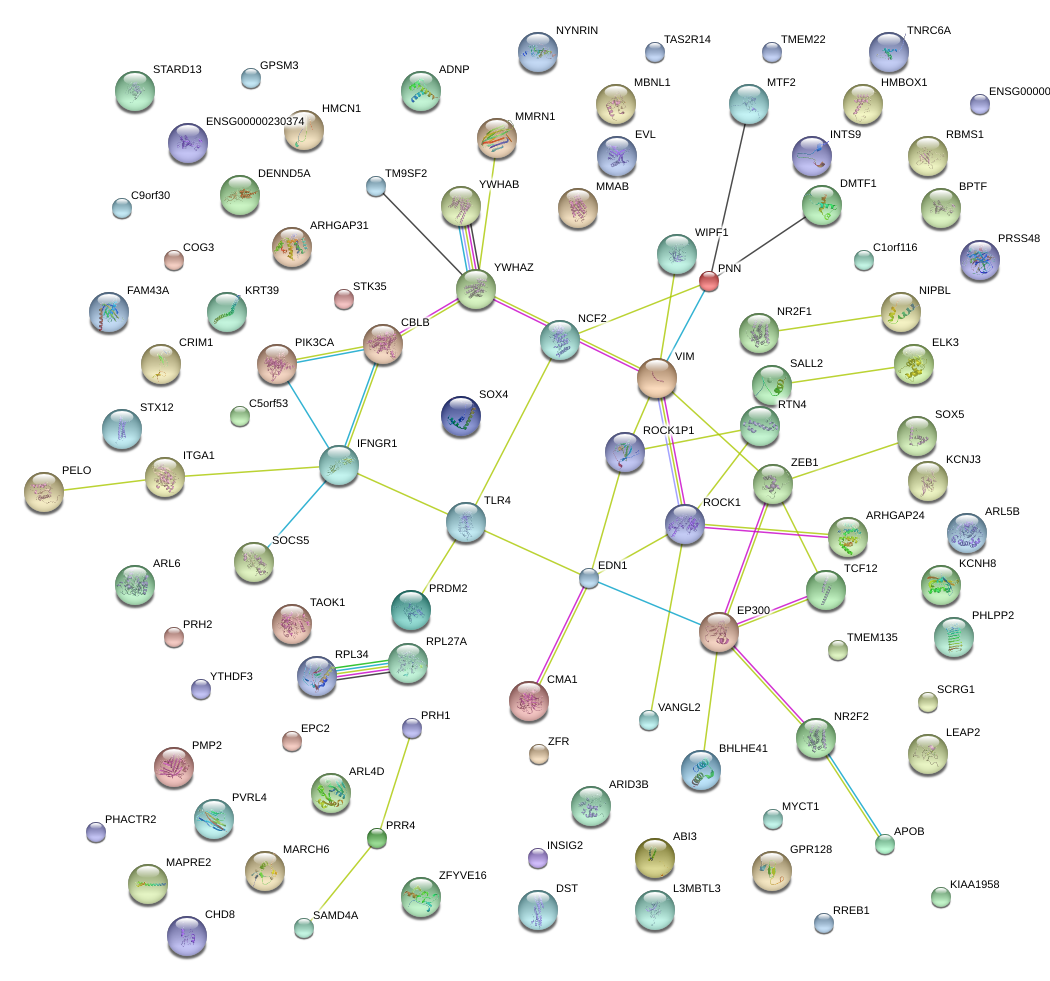
|
chr18_+_30875297
|
2.809
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr3_+_180348818
|
1.952
|
NM_006218
|
PIK3CA
|
phosphoinositide-3-kinase, catalytic, alpha polypeptide
|
|
chr6_+_130381408
|
1.486
|
NM_001007102
NM_032438
|
L3MBTL3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr5_+_139485703
|
1.316
|
NM_001007189
|
C5orf53
|
chromosome 5 open reading frame 53
|
|
chr13_-_32678142
|
1.114
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr11_+_86426518
|
1.095
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr2_+_36436873
|
0.950
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr4_+_152417774
|
0.945
|
NM_183375
|
PRSS48
|
protease, serine, 48
|
|
chr6_+_12398514
|
0.935
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr8_+_64243741
|
0.907
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr8_+_64243662
|
0.904
|
NM_152758
|
YTHDF3
|
YTH domain family, member 3
|
|
chr6_+_144040751
|
0.876
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr6_+_7052798
|
0.848
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr14_+_99601498
|
0.844
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr20_-_48980858
|
0.841
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr1_-_205272619
|
0.780
|
NM_001083924
NM_023938
|
C1orf116
|
chromosome 1 open reading frame 116
|
|
chr1_+_158636987
|
0.767
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr7_-_104440707
|
0.765
|
|
LOC100216545
|
hypothetical LOC100216545
|
|
chr6_+_7053183
|
0.732
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr20_-_2031521
|
0.727
|
|
|
|
|
chr8_+_28803804
|
0.724
|
NM_001135726
|
HMBOX1
|
homeobox containing 1
|
|
chr2_-_175207521
|
0.715
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr2_-_21084911
|
0.712
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr14_+_99601517
|
0.711
|
|
EVL
|
Enah/Vasp-like
|
|
chr12_-_26166485
|
0.703
|
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr16_+_24696069
|
0.690
|
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr11_-_9182356
|
0.690
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr3_+_153499883
|
0.690
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr8_-_28803341
|
0.689
|
NM_001172562
|
INTS9
|
integrator complex subunit 9
|
|
chr8_-_28803616
|
0.670
|
NM_001145159
NM_018250
|
INTS9
|
integrator complex subunit 9
|
|
chr17_+_24742068
|
0.670
|
NM_020791
|
TAOK1
|
TAO kinase 1
|
|
chr3_+_120495906
|
0.665
|
NM_020754
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr4_+_87070449
|
0.628
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_+_27972269
|
0.627
|
NM_177424
|
STX12
|
syntaxin 12
|
|
chr20_+_2031418
|
0.614
|
|
STK35
|
serine/threonine kinase 35
|
|
chr5_+_10406829
|
0.611
|
|
MARCH6
|
membrane-associated ring finger (C3HC4) 6
|
|
chr6_+_7052985
|
0.608
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr2_+_149118791
|
0.606
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr2_+_155263338
|
0.606
|
NM_002239
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3
|
|
chr14_+_38720085
|
0.605
|
|
PNN
|
pinin, desmosome associated protein
|
|
chr17_+_44643067
|
0.604
|
|
ABI3
|
ABI family, member 3
|
|
chr16_+_31105579
|
0.599
|
|
|
|
|
chr9_+_119506446
|
0.597
|
|
TLR4
|
toll-like receptor 4
|
|
chr9_+_119506274
|
0.592
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr12_-_70953461
|
0.590
|
|
LOC283392
|
hypothetical LOC283392
|
|
chr3_+_138020550
|
0.579
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr12_-_11215396
|
0.579
|
NM_001098538
NM_006250
|
PRH1-PRR4
PRR4
TAS2R14
PRH1
|
PRH1-PRR4 read-through transcript
proline rich 4 (lacrimal)
taste receptor, type 2, member 14
proline-rich protein HaeIII subfamily 1
|
|
chr14_-_20969706
|
0.572
|
NM_001170629
|
CHD8
|
chromodomain helicase DNA binding protein 8
|
|
chr2_+_46779553
|
0.561
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr3_+_98966253
|
0.555
|
NM_032146
NM_177976
|
ARL6
|
ADP-ribosylation factor-like 6
|
|
chr15_+_94676691
|
0.551
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr17_-_36376663
|
0.550
|
NM_213656
|
KRT39
|
keratin 39
|
|
chr13_+_44937025
|
0.549
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr9_+_114289178
|
0.546
|
|
KIAA1958
|
KIAA1958
|
|
chr9_+_119506428
|
0.542
|
|
TLR4
|
toll-like receptor 4
|
|
chr6_-_56816403
|
0.540
|
|
DST
|
dystonin
|
|
chr15_+_55298908
|
0.522
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr1_-_181826633
|
0.521
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr5_+_52131686
|
0.515
|
|
PELO
|
pelota homolog (Drosophila)
|
|
chr3_+_101811122
|
0.509
|
NM_032787
|
GPR128
|
G protein-coupled receptor 128
|
|
chr5_-_32439067
|
0.505
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr3_+_195889845
|
0.502
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr12_+_95112385
|
0.501
|
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr6_+_21701942
|
0.497
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr10_+_31647425
|
0.496
|
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr6_+_153060722
|
0.496
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr2_-_160838397
|
0.491
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr17_+_63252704
|
0.491
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr22_+_39818553
|
0.490
|
NM_001429
|
EP300
|
E1A binding protein p300
|
|
chr5_+_132237253
|
0.489
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr18_-_16945763
|
0.486
|
NM_005406
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr12_-_23995109
|
0.485
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr11_+_8660913
|
0.483
|
|
RPL27A
|
ribosomal protein L27a
|
|
chr14_-_21074875
|
0.479
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr4_+_91035025
|
0.477
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr2_+_118562499
|
0.476
|
NM_016133
|
INSIG2
|
insulin induced gene 2
|
|
chr20_+_42947754
|
0.476
|
NM_003404
NM_139323
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr14_+_54104099
|
0.472
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr7_+_86619580
|
0.472
|
NM_001142326
NM_001142327
|
DMTF1
|
cyclin D binding myb-like transcription factor 1
|
|
chr20_+_42947764
|
0.471
|
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr1_+_93317374
|
0.471
|
NM_001164391
NM_001164392
NM_001164393
NM_007358
|
MTF2
|
metal response element binding transcription factor 2
|
|
chr15_+_72620570
|
0.471
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr4_-_174557191
|
0.469
|
NM_007281
|
SCRG1
|
stimulator of chondrogenesis 1
|
|
chr2_-_55090759
|
0.466
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr1_+_13948448
|
0.464
|
NM_001007257
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr13_+_98951772
|
0.463
|
|
TM9SF2
|
transmembrane 9 superfamily member 2
|
|
chr5_+_52119442
|
0.459
|
NM_181501
NM_015946
|
ITGA1
PELO
|
integrin, alpha 1
pelota homolog (Drosophila)
|
|
chr11_+_64950074
|
0.453
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr5_+_36912656
|
0.452
|
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr10_+_17312654
|
0.450
|
|
VIM
|
vimentin
|
|
chr4_+_109761170
|
0.449
|
NM_000995
NM_033625
|
RPL34
|
ribosomal protein L34
|
|
chr9_+_102244005
|
0.445
|
NM_001198812
|
C9orf3-TMEFF1
C9orf30
|
C9orf3-TMEFF1 readthrough
chromosome 9 open reading frame 30
|
|
chr1_+_183970093
|
0.440
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr16_-_70306198
|
0.438
|
NM_015020
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2
|
|
chr14_-_24047261
|
0.437
|
NM_001836
|
CMA1
|
chymase 1, mast cell
|
|
chr14_+_23937766
|
0.435
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr5_+_79739548
|
0.435
|
NM_001105251
NM_014733
|
ZFYVE16
|
zinc finger, FYVE domain containing 16
|
|
chr8_-_82522175
|
0.434
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr2_+_36436316
|
0.433
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr6_-_32268267
|
0.432
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr3_+_120496180
|
0.431
|
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr10_+_18988282
|
0.430
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr10_-_18988172
|
0.430
|
|
|
|
|
chr3_-_107070400
|
0.426
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr5_-_58370693
|
0.422
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr10_-_27189898
|
0.421
|
|
ABI1
|
abl-interactor 1
|
|
chr6_+_7053863
|
0.420
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr14_+_49157150
|
0.419
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr2_+_149119029
|
0.419
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr8_+_90983934
|
0.411
|
NM_001126111
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2
|
|
chr20_+_13924263
|
0.406
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr2_+_64604940
|
0.406
|
NM_001002243
NM_017657
NM_203437
|
AFTPH
|
aftiphilin
|
|
chr2_+_54638957
|
0.404
|
NM_178313
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr3_+_114948550
|
0.403
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr20_+_30409649
|
0.402
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr8_-_38245694
|
0.402
|
NM_001102559
NM_001102560
NM_032483
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr11_-_5419358
|
0.400
|
NM_001005288
|
OR51I1
|
olfactory receptor, family 51, subfamily I, member 1
|
|
chr1_+_36169260
|
0.399
|
NM_024852
NM_177422
|
EIF2C3
|
eukaryotic translation initiation factor 2C, 3
|
|
chr14_+_64041170
|
0.398
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr13_+_98951653
|
0.396
|
NM_004800
|
TM9SF2
|
transmembrane 9 superfamily member 2
|
|
chr21_-_30614477
|
0.394
|
NM_203405
|
KRTAP26-1
|
keratin associated protein 26-1
|
|
chr5_-_54504590
|
0.393
|
NM_001145734
NM_001170402
NM_152623
|
CDC20B
|
cell division cycle 20 homolog B (S. cerevisiae)
|
|
chr17_+_63252668
|
0.393
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr6_+_47732181
|
0.392
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr12_-_15005761
|
0.390
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr3_-_69674412
|
0.390
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr22_-_34566210
|
0.388
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr2_+_187162969
|
0.387
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr12_+_51686512
|
0.386
|
|
EIF4B
|
eukaryotic translation initiation factor 4B
|
|
chr2_-_47986160
|
0.384
|
NM_001190274
|
FBXO11
|
F-box protein 11
|
|
chr1_+_246635918
|
0.384
|
NM_030904
|
OR2T1
|
olfactory receptor, family 2, subfamily T, member 1
|
|
chr22_-_34566367
|
0.384
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr3_+_99934227
|
0.384
|
NM_006100
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
|
chr20_-_42249631
|
0.383
|
NM_020433
NM_175913
|
JPH2
|
junctophilin 2
|
|
chr2_-_213723298
|
0.381
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr5_+_112340326
|
0.380
|
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chr16_-_65781499
|
0.379
|
NM_178516
|
EXOC3L1
|
exocyst complex component 3-like 1
|
|
chr11_+_64950402
|
0.379
|
|
|
|
|
chr9_+_114288899
|
0.375
|
NM_133465
|
KIAA1958
|
KIAA1958
|
|
chr1_+_182040837
|
0.375
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr1_+_26896519
|
0.374
|
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr6_+_57290357
|
0.371
|
NM_000947
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa)
|
|
chr22_-_34566276
|
0.370
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr3_-_114176401
|
0.370
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr7_+_37926943
|
0.369
|
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr3_+_69216747
|
0.368
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chr20_+_8997660
|
0.364
|
NM_001172646
|
PLCB4
|
phospholipase C, beta 4
|
|
chr9_+_35528890
|
0.363
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr4_+_77575276
|
0.361
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr15_-_56093477
|
0.360
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr18_-_16945613
|
0.360
|
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr13_+_92677012
|
0.359
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr5_+_10406802
|
0.359
|
NM_005885
|
MARCH6
|
membrane-associated ring finger (C3HC4) 6
|
|
chr10_+_116843108
|
0.358
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr5_+_10406980
|
0.352
|
|
|
|
|
chr12_+_102505180
|
0.351
|
NM_017564
|
STAB2
|
stabilin 2
|
|
chr16_+_24475129
|
0.351
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr6_-_11490487
|
0.350
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr1_+_152229861
|
0.349
|
NM_001030
|
RPS27
|
ribosomal protein S27
|
|
chr5_+_65258250
|
0.347
|
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr12_+_45896511
|
0.345
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr8_-_88955340
|
0.344
|
NM_152418
|
DCAF4L2
|
DDB1 and CUL4 associated factor 4-like 2
|
|
chr1_+_22224571
|
0.343
|
|
HSPC157
|
hypothetical LOC29092
|
|
chr5_+_65258139
|
0.341
|
NM_001006600
NM_018695
|
ERBB2IP
|
erbb2 interacting protein
|
|
chr14_+_64040979
|
0.341
|
NM_001123329
NM_014950
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr9_+_108665168
|
0.338
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr10_+_37454790
|
0.337
|
NM_052997
|
ANKRD30A
|
ankyrin repeat domain 30A
|
|
chr6_+_18495559
|
0.336
|
NM_182757
|
RNF144B
|
ring finger protein 144B
|
|
chr12_-_26877189
|
0.334
|
NM_002223
|
ITPR2
|
inositol 1,4,5-triphosphate receptor, type 2
|
|
chr8_+_9990471
|
0.333
|
NM_001135671
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr2_+_101874765
|
0.332
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr5_+_67558217
|
0.332
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr2_-_202811459
|
0.331
|
NM_001005781
NM_001005782
NM_003352
|
SUMO1
|
SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae)
|
|
chr12_-_53071285
|
0.331
|
NM_001130967
NM_001130968
|
ZNF385A
|
zinc finger protein 385A
|
|
chr10_+_106003941
|
0.330
|
NM_001191003
|
GSTO1
|
glutathione S-transferase omega 1
|
|
chr3_-_158695832
|
0.329
|
NM_001167917
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr5_+_173247919
|
0.327
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr4_-_151993422
|
0.326
|
|
|
|
|
chr11_+_22646229
|
0.325
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr8_+_7789608
|
0.324
|
NM_004942
|
DEFB4A
|
defensin, beta 4A
|
|
chr2_-_51113040
|
0.324
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr7_+_138566770
|
0.324
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr5_-_10406690
|
0.324
|
|
|
|
|
chr1_+_25629968
|
0.323
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr4_-_103968104
|
0.322
|
NM_003340
NM_181886
NM_181888
NM_181889
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast)
|
|
chr1_+_64983312
|
0.322
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr17_+_55139792
|
0.320
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr17_+_55139807
|
0.320
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr3_+_197951203
|
0.319
|
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr15_-_54175954
|
0.317
|
|
RFX7
|
regulatory factor X, 7
|
|
chr1_-_177378801
|
0.317
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr2_-_51112743
|
0.317
|
|
NRXN1
|
neurexin 1
|
|
chr17_-_55139538
|
0.316
|
NM_016077
|
PTRH2
|
peptidyl-tRNA hydrolase 2
|
|
chr17_+_55139743
|
0.316
|
|
VMP1
|
vacuole membrane protein 1
|
|
chr1_+_66772393
|
0.316
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr2_-_202192097
|
0.316
|
|
ALS2CR11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11
|
|
chr7_+_128651976
|
0.315
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr3_+_87359088
|
0.314
|
NM_014043
|
CHMP2B
|
chromatin modifying protein 2B
|
|
chr2_+_201384505
|
0.314
|
|
BZW1
|
basic leucine zipper and W2 domains 1
|
|
chr14_+_64041257
|
0.314
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|