|
chr19_-_10625449
|
3.727
|
|
LOC147727
|
hypothetical LOC147727
|
|
chr19_+_10625987
|
3.487
|
|
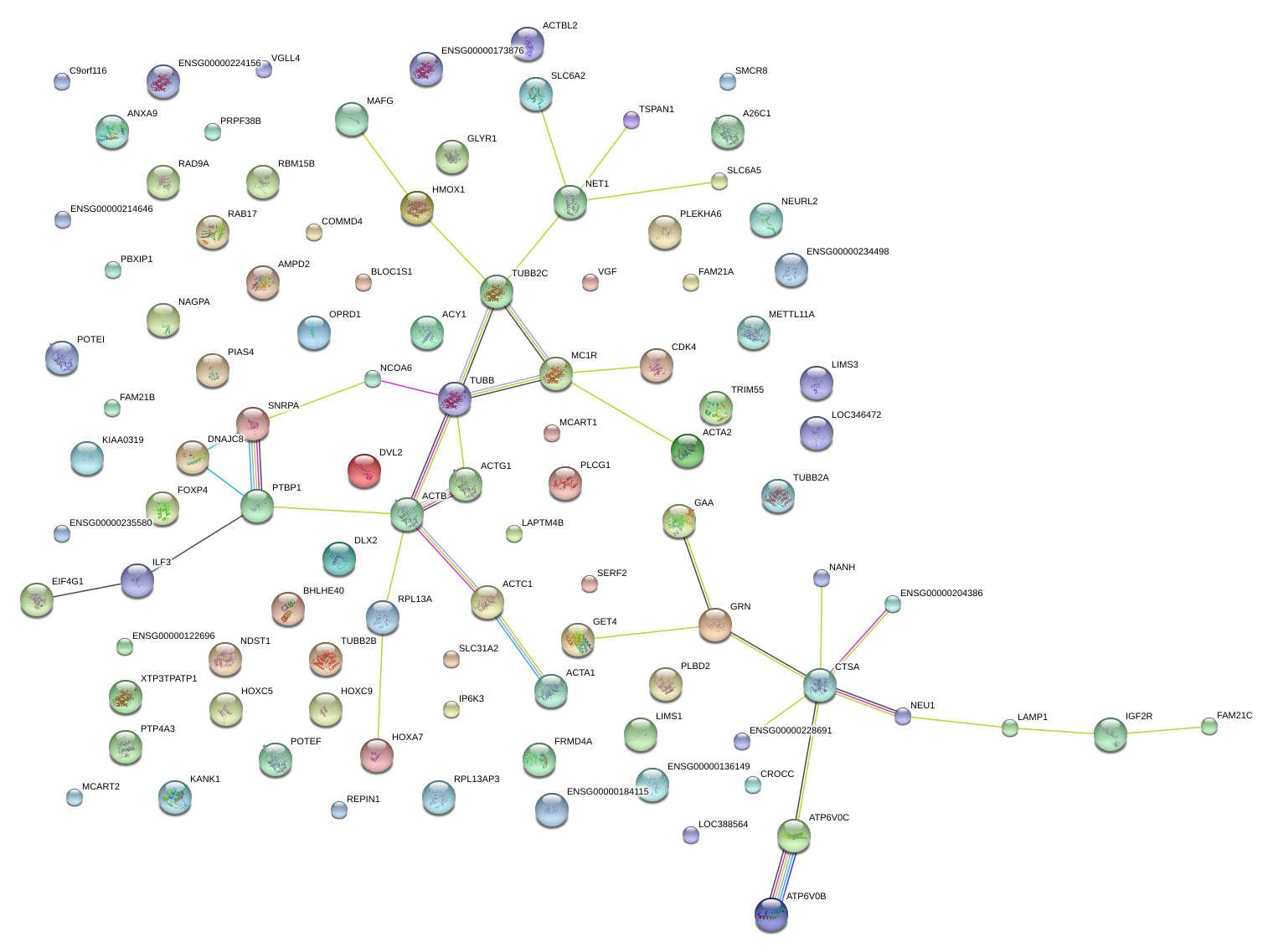
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr19_+_10625896
|
3.262
|
NM_001137673
NM_004516
NM_012218
NM_017620
NM_153464
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr19_+_10626106
|
3.050
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr20_+_39199542
|
1.573
|
NM_002660
NM_182811
|
PLCG1
|
phospholipase C, gamma 1
|
|
chr19_+_748390
|
1.496
|
NM_002819
NM_031990
NM_031991
NM_175847
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr19_+_748444
|
1.492
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr20_-_39199936
|
1.441
|
|
|
|
|
chr7_+_882704
|
1.407
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr17_-_7078399
|
1.345
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr17_-_7078304
|
1.293
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr17_-_7078446
|
1.266
|
NM_004422
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr19_+_3958690
|
1.262
|
NM_015897
|
PIAS4
|
protein inhibitor of activated STAT, 4
|
|
chr7_+_882766
|
1.259
|
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr10_+_5478521
|
1.209
|
NM_005863
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr10_-_14090459
|
1.199
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr20_+_43953428
|
1.101
|
|
CTSA
|
cathepsin A
|
|
chr20_+_43953606
|
1.101
|
|
CTSA
|
cathepsin A
|
|
chr17_+_75851984
|
1.098
|
|
|
|
|
chr7_+_149696792
|
1.044
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr12_-_56432375
|
1.036
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_+_52713098
|
1.033
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr17_+_77478858
|
1.031
|
|
LOC92659
|
hypothetical LOC92659
|
|
chr12_-_56432377
|
0.998
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432292
|
0.988
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr17_-_77478835
|
0.966
|
NM_002359
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr12_-_56432385
|
0.966
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr19_+_54682661
|
0.958
|
|
RPL13A
|
ribosomal protein L13a
|
|
chr1_+_44213200
|
0.957
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr3_+_4996096
|
0.950
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr6_-_169392771
|
0.941
|
|
|
|
|
chr17_+_39778016
|
0.939
|
NM_002087
|
GRN
|
granulin
|
|
chr22_+_34107087
|
0.936
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr2_-_172675873
|
0.930
|
|
DLX2
|
distal-less homeobox 2
|
|
chr17_+_75689985
|
0.927
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr6_+_160310118
|
0.907
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr1_+_44213210
|
0.893
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr1_+_44213229
|
0.891
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr11_+_66915997
|
0.887
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr1_+_17121012
|
0.880
|
NM_014675
|
CROCC
|
ciliary rootlet coiled-coil, rootletin
|
|
chr6_-_33822659
|
0.871
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr9_-_37894098
|
0.866
|
|
MCART1
|
mitochondrial carrier triple repeat 1
|
|
chr19_+_45948598
|
0.862
|
NM_004596
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A
|
|
chr12_+_54396099
|
0.857
|
|
BLOC1S1-RDH5
BLOC1S1
|
BLOC1S1-RDH5 read-through transcript
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr1_+_44213260
|
0.854
|
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr1_+_44213182
|
0.845
|
NM_001039457
NM_004047
|
ATP6V0B
|
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b
|
|
chr20_+_43953331
|
0.842
|
|
CTSA
|
cathepsin A
|
|
chr12_+_112280753
|
0.831
|
NM_001159727
NM_173542
|
PLBD2
|
phospholipase B domain containing 2
|
|
chr17_+_18159333
|
0.811
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr3_+_51403760
|
0.811
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr12_+_54396086
|
0.810
|
NM_001487
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr1_-_28432060
|
0.808
|
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr9_-_137531517
|
0.798
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr20_+_43953372
|
0.792
|
|
CTSA
|
cathepsin A
|
|
chr6_+_30796064
|
0.785
|
|
TUBB
|
tubulin, beta
|
|
chr8_+_67201684
|
0.781
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr17_+_75689894
|
0.781
|
NM_000152
NM_001079803
NM_001079804
|
GAA
|
glucosidase, alpha; acid
|
|
chr3_-_11585264
|
0.777
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr19_+_748452
|
0.776
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr16_-_4837266
|
0.775
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr16_-_5023934
|
0.768
|
NM_016256
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr7_+_149696901
|
0.759
|
|
REPIN1
|
replication initiator 1
|
|
chr1_+_109963982
|
0.758
|
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr16_-_5023914
|
0.744
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr3_+_4996141
|
0.740
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr6_+_33045070
|
0.738
|
|
|
|
|
chr1_-_28432095
|
0.737
|
NM_014280
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr17_+_75689955
|
0.734
|
|
GAA
|
glucosidase, alpha; acid
|
|
chr8_+_142501188
|
0.732
|
NM_007079
NM_032611
|
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3
|
|
chr12_+_54396118
|
0.732
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr19_+_748448
|
0.731
|
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chr10_+_45542653
|
0.727
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr19_+_54682676
|
0.724
|
NM_012423
|
RPL13A
|
ribosomal protein L13a
|
|
chr1_+_29011240
|
0.716
|
NM_000911
|
OPRD1
|
opioid receptor, delta 1
|
|
chr1_-_153195188
|
0.713
|
NM_020524
|
PBXIP1
|
pre-B-cell leukemia homeobox interacting protein 1
|
|
chr8_+_98857190
|
0.711
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr2_-_238164042
|
0.710
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr17_+_18159330
|
0.699
|
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr7_-_27162688
|
0.697
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr17_+_18159294
|
0.692
|
NM_144775
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8
|
|
chr3_+_185515013
|
0.689
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr9_+_131428272
|
0.684
|
|
METTL11A
|
methyltransferase like 11A
|
|
chr17_-_77092569
|
0.683
|
|
ACTG1
|
actin, gamma 1
|
|
chr17_+_39778191
|
0.682
|
|
GRN
|
granulin
|
|
chr13_+_112999538
|
0.681
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr17_+_39778153
|
0.676
|
|
GRN
|
granulin
|
|
chr17_+_39778180
|
0.674
|
|
GRN
|
granulin
|
|
chr7_-_100595552
|
0.674
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr22_+_34107051
|
0.671
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr3_+_51992339
|
0.667
|
NM_000666
NM_001198895
NM_001198896
NM_001198897
NM_001198898
|
ACY1
|
aminoacylase 1
|
|
chr1_-_202595666
|
0.665
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr6_+_30796123
|
0.663
|
NM_178014
|
TUBB
|
tubulin, beta
|
|
chr15_+_41871768
|
0.663
|
NM_001018108
|
SERF2
|
small EDRK-rich factor 2
|
|
chr12_+_52680143
|
0.662
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr19_-_60587414
|
0.661
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr9_+_696744
|
0.660
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr1_+_109963950
|
0.659
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr5_+_149845503
|
0.655
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr16_+_2503978
|
0.652
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr16_+_2503915
|
0.647
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr16_+_2503994
|
0.641
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr16_+_5023672
|
0.640
|
|
LOC100507589
|
hypothetical LOC100507589
|
|
chr2_+_108571336
|
0.639
|
NM_001193488
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr6_+_30796121
|
0.636
|
|
TUBB
|
tubulin, beta
|
|
chr6_+_41622111
|
0.636
|
NM_001012426
NM_001012427
NM_138457
|
FOXP4
|
forkhead box P4
|
|
chr1_+_224337937
|
0.635
|
|
|
|
|
chr2_+_108571209
|
0.634
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr9_+_131428225
|
0.626
|
NM_014064
|
METTL11A
|
methyltransferase like 11A
|
|
chr9_+_114953048
|
0.623
|
NM_001860
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr9_+_114953087
|
0.622
|
|
SLC31A2
|
solute carrier family 31 (copper transporters), member 2
|
|
chr6_-_31938530
|
0.617
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr16_+_2503676
|
0.617
|
NM_001198569
NM_001694
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr1_+_149221122
|
0.617
|
NM_003568
|
ANXA9
|
annexin A9
|
|
chr15_+_73415426
|
0.616
|
NM_017828
|
COMMD4
|
COMM domain containing 4
|
|
chr3_+_185515108
|
0.612
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr20_-_43953036
|
0.609
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr6_+_33044353
|
0.609
|
|
|
|
|
chr6_+_33044378
|
0.600
|
NM_005104
|
BRD2
|
bromodomain containing 2
|
|
chr20_+_33823318
|
0.598
|
NM_016436
|
PHF20
|
PHD finger protein 20
|
|
chr16_-_5023562
|
0.591
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr22_+_38903825
|
0.590
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr6_+_151815114
|
0.586
|
NM_024573
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr2_+_108571177
|
0.583
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr3_+_4996305
|
0.581
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr8_+_98856984
|
0.578
|
NM_018407
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr16_+_11669736
|
0.576
|
NM_003498
|
SNN
|
stannin
|
|
chr17_-_44043361
|
0.574
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr20_-_60484420
|
0.566
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chr19_-_13088212
|
0.564
|
NM_001142554
NM_017722
NM_001136035
|
TRMT1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr20_-_61317891
|
0.557
|
NM_017798
|
YTHDF1
|
YTH domain family, member 1
|
|
chr15_+_41871809
|
0.553
|
|
SERF2-C15ORF63
SERF2
|
SERF2-C15orf63 read-through transcript
small EDRK-rich factor 2
|
|
chr16_+_2504001
|
0.550
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr16_+_22124870
|
0.542
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr4_+_17225344
|
0.541
|
NM_025205
|
MED28
|
mediator complex subunit 28
|
|
chr17_-_44043278
|
0.537
|
|
HOXB7
|
homeobox B7
|
|
chr13_+_112999590
|
0.528
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr16_+_66676833
|
0.519
|
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr17_-_18158928
|
0.518
|
NM_004618
|
TOP3A
|
topoisomerase (DNA) III alpha
|
|
chr16_+_66676851
|
0.517
|
NM_004555
NM_173163
NM_173164
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr16_-_66072477
|
0.517
|
NM_004691
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1
|
|
chr12_-_9913199
|
0.516
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr5_+_244625
|
0.515
|
NM_001080478
|
LRRC14B
|
leucine rich repeat containing 14B
|
|
chr14_-_93919305
|
0.515
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr10_+_101532450
|
0.514
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr1_+_11788827
|
0.512
|
|
CLCN6
|
chloride channel 6
|
|
chr20_+_43028533
|
0.512
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr10_+_18988282
|
0.509
|
NM_178815
|
ARL5B
|
ADP-ribosylation factor-like 5B
|
|
chr7_-_1562272
|
0.506
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr8_+_142471290
|
0.505
|
|
|
|
|
chr17_+_22645232
|
0.503
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr15_-_48766018
|
0.502
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr7_+_142723340
|
0.500
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr19_-_45546250
|
0.497
|
|
C19orf47
|
chromosome 19 open reading frame 47
|
|
chr1_+_110683580
|
0.497
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr16_-_71549149
|
0.496
|
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr1_+_3360880
|
0.496
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr6_-_33493640
|
0.495
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr9_-_133574928
|
0.493
|
NM_198679
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr1_+_110683586
|
0.492
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr2_+_26840626
|
0.490
|
NM_017877
|
CENPA
C2orf18
|
centromere protein A
chromosome 2 open reading frame 18
|
|
chr10_+_101532544
|
0.487
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr22_+_21318779
|
0.485
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr8_-_143482433
|
0.481
|
NM_145003
|
TSNARE1
|
t-SNARE domain containing 1
|
|
chr1_-_159459999
|
0.480
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr19_-_45948211
|
0.480
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr7_-_27108826
|
0.476
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr16_+_29983216
|
0.476
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr1_+_110683467
|
0.473
|
NM_022768
|
RBM15
|
RNA binding motif protein 15
|
|
chr14_+_19993181
|
0.470
|
|
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1
|
|
chr17_-_44042924
|
0.469
|
|
HOXB7
|
homeobox B7
|
|
chr22_+_36928972
|
0.466
|
NM_001161573
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr10_-_18988172
|
0.464
|
|
|
|
|
chr5_-_176663265
|
0.458
|
NM_001031677
NM_130781
|
RAB24
|
RAB24, member RAS oncogene family
|
|
chr6_+_36670067
|
0.457
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr6_+_138230045
|
0.455
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr2_+_223625105
|
0.455
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr2_+_74914742
|
0.453
|
|
HK2
|
hexokinase 2
|
|
chr20_+_43028590
|
0.445
|
|
STK4
|
serine/threonine kinase 4
|
|
chr17_-_44022595
|
0.445
|
|
HOXB3
|
homeobox B3
|
|
chr19_+_1358750
|
0.444
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr14_-_63830830
|
0.443
|
NM_001040275
NM_001437
|
ESR2
|
estrogen receptor 2 (ER beta)
|
|
chr6_+_35852369
|
0.442
|
NM_207409
|
C6orf126
|
chromosome 6 open reading frame 126
|
|
chr17_-_46692369
|
0.440
|
NM_017643
|
MBTD1
|
mbt domain containing 1
|
|
chr16_+_29983473
|
0.440
|
NM_001127617
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr17_+_34610097
|
0.438
|
|
RPL19
|
ribosomal protein L19
|
|
chr9_+_6747636
|
0.436
|
NM_001146694
NM_001146695
NM_015061
|
KDM4C
|
lysine (K)-specific demethylase 4C
|
|
chr12_+_6514645
|
0.436
|
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase
|
|
chr17_-_41018291
|
0.436
|
|
|
|
|
chr16_+_4837912
|
0.435
|
NM_001079514
|
UBN1
|
ubinuclein 1
|
|
chr2_+_215885063
|
0.434
|
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
|
|
chr5_+_134012133
|
0.433
|
NM_021982
|
SEC24A
|
SEC24 family, member A (S. cerevisiae)
|
|
chr6_+_36670117
|
0.431
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr14_-_19993032
|
0.430
|
NM_017807
|
OSGEP
|
O-sialoglycoprotein endopeptidase
|
|
chr1_+_10015577
|
0.429
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chr1_+_32992131
|
0.428
|
|
|
|
|
chr8_+_17824631
|
0.428
|
NM_006197
|
PCM1
|
pericentriolar material 1
|
|
chr17_+_6856521
|
0.426
|
NM_001004333
|
RNASEK-C17ORF49
RNASEK
|
RNASEK-C17orf49 read-through transcript
ribonuclease, RNase K
|
|
chr12_+_51686504
|
0.424
|
|
EIF4B
|
eukaryotic translation initiation factor 4B
|
|
chr19_-_45948189
|
0.423
|
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr3_-_188946168
|
0.423
|
NM_001706
|
BCL6
|
B-cell CLL/lymphoma 6
|