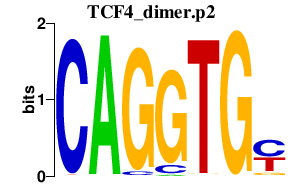
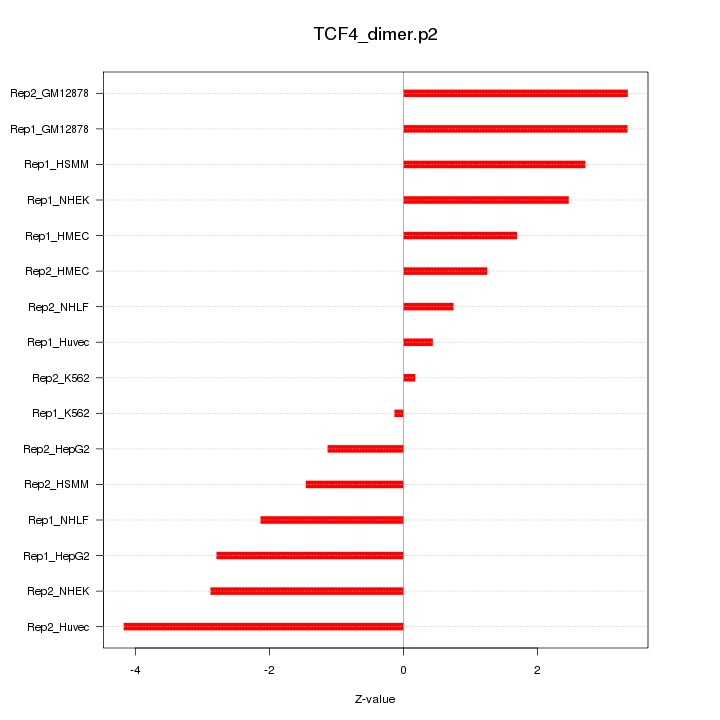
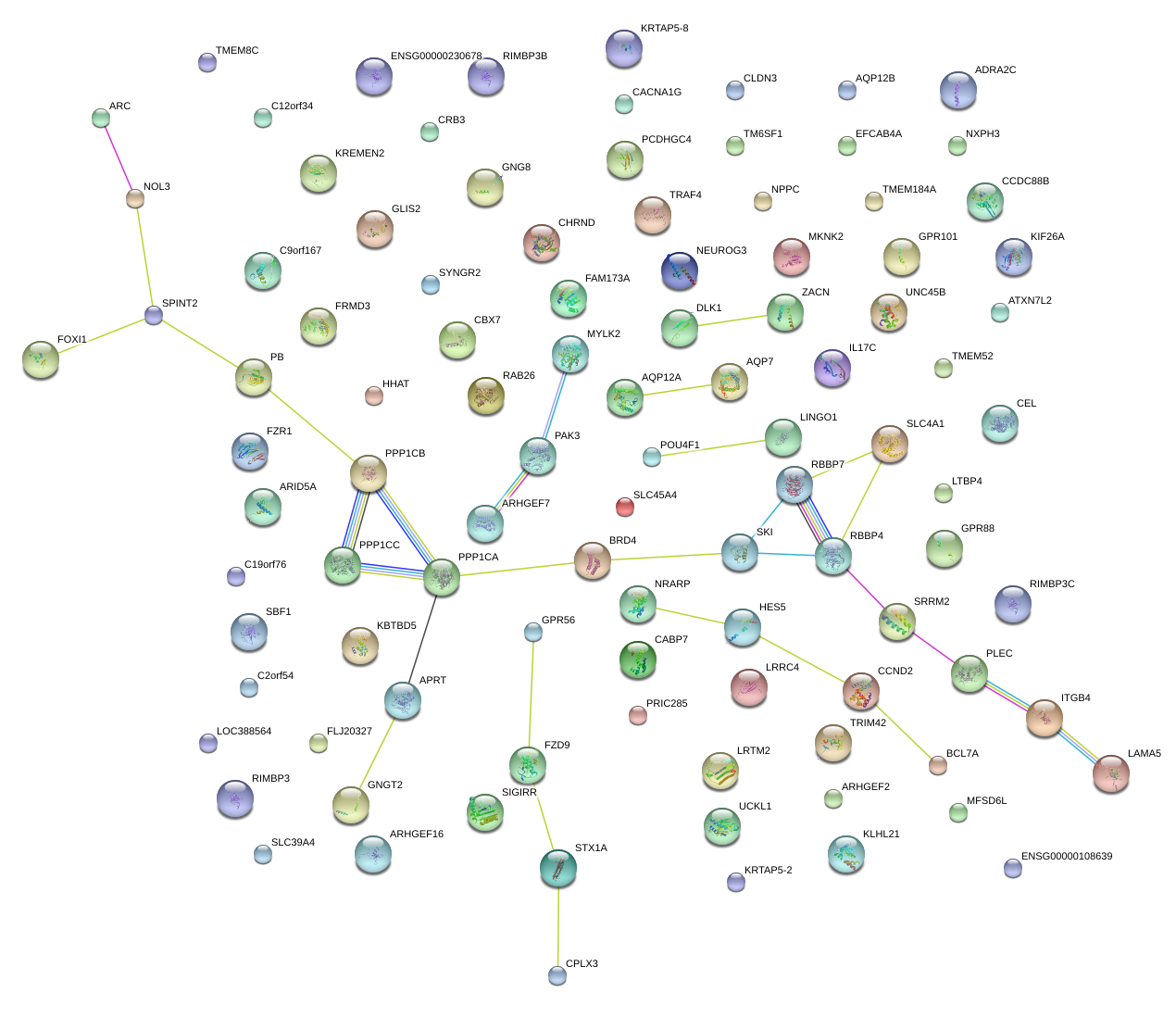
|
chr17_+_30498948
|
3.660
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr11_+_63864263
|
3.166
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr22_-_49240594
|
2.885
|
|
SBF1
|
SET binding factor 1
|
|
chr9_+_139292094
|
2.808
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr11_+_817584
|
2.765
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr2_-_241484245
|
2.761
|
NM_001085437
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr20_-_61669550
|
2.744
|
NM_033405
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chrX_-_135941498
|
2.697
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr22_+_20068039
|
2.574
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr11_+_70926718
|
2.515
|
NM_021046
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr1_-_1840564
|
2.514
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr1_+_3360880
|
2.449
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr16_+_87232501
|
2.443
|
NM_013278
|
IL17C
|
interleukin 17C
|
|
chr16_-_87405817
|
2.425
|
NM_000485
NM_001030018
|
APRT
|
adenine phosphoribosyltransferase
|
|
chr8_-_145088679
|
2.357
|
NM_201383
|
PLEC
|
plectin
|
|
chr17_+_24095135
|
2.333
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chrX_+_110226030
|
2.241
|
NM_002578
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr8_-_145609737
|
2.139
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr17_+_26060617
|
2.135
|
|
|
|
|
chr12_+_120944174
|
2.135
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr17_-_8643163
|
2.132
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr15_-_75694674
|
2.045
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr2_-_241270982
|
2.024
|
NM_001102467
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr16_+_56230707
|
2.015
|
NM_001145774
|
GPR56
|
G protein-coupled receptor 56
|
|
chr5_+_169465478
|
1.926
|
NM_012188
NM_144769
|
FOXI1
|
forkhead box I1
|
|
chr3_+_42702014
|
1.914
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr19_+_54883732
|
1.879
|
NM_001101340
|
C19orf76
|
chromosome 19 open reading frame 76
|
|
chr1_-_6585268
|
1.852
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr1_-_154205675
|
1.847
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr19_+_45799060
|
1.840
|
NM_001042545
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr9_+_134927185
|
1.840
|
NM_001807
|
CEL
|
carboxyl ester lipase (bile salt-stimulated lipase)
|
|
chr7_+_72486044
|
1.827
|
NM_003508
|
FZD9
|
frizzled homolog 9 (Drosophila)
|
|
chr7_-_1562272
|
1.808
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr7_-_72822471
|
1.800
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr9_-_135379888
|
1.776
|
NM_001080483
|
TMEM8C
|
transmembrane protein 8C
|
|
chr14_-_105392671
|
1.772
|
|
|
|
|
chr1_+_1062137
|
1.770
|
|
LOC254099
|
hypothetical protein LOC254099
|
|
chr17_-_39700960
|
1.754
|
NM_000342
|
SLC4A1
|
solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group)
|
|
chr19_-_15252261
|
1.740
|
NM_014299
NM_058243
|
BRD4
|
bromodomain containing 4
|
|
chr16_+_711139
|
1.728
|
NM_023933
|
FAM173A
|
family with sequence similarity 173, member A
|
|
chr19_-_60587414
|
1.725
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr8_-_143692814
|
1.721
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr20_-_62052922
|
1.719
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr12_+_108690199
|
1.718
|
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr13_-_78075683
|
1.710
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr16_+_4325142
|
1.700
|
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr20_+_29870836
|
1.677
|
NM_033118
|
MYLK2
|
myosin light chain kinase 2
|
|
chr22_+_28446288
|
1.669
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr19_+_3457264
|
1.655
|
NM_016263
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr17_+_71235324
|
1.647
|
|
ITGB4
|
integrin, beta 4
|
|
chr5_+_140844810
|
1.639
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr20_-_60375695
|
1.638
|
NM_005560
|
LAMA5
|
laminin, alpha 5
|
|
chr19_+_6415259
|
1.635
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr11_-_66945190
|
1.623
|
|
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme
|
|
chr14_+_100270222
|
1.622
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr12_+_4253143
|
1.614
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr1_+_2149993
|
1.600
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr22_-_20235372
|
1.593
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr19_+_43447262
|
1.587
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr2_+_233099113
|
1.585
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr10_-_71002998
|
1.576
|
NM_020999
|
NEUROG3
|
neurogenin 3
|
|
chr11_-_407324
|
1.573
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr19_-_51829778
|
1.570
|
NM_033258
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8
|
|
chr16_+_2954217
|
1.552
|
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr19_+_6723801
|
1.522
|
|
|
|
|
chr7_-_72761367
|
1.515
|
|
STX1A
|
syntaxin 1A (brain)
|
|
chr16_+_2138688
|
1.514
|
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr12_+_1799919
|
1.514
|
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr3_+_141879555
|
1.511
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr19_+_43446937
|
1.507
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr16_+_2138608
|
1.505
|
NM_014353
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr17_+_71586857
|
1.503
|
NM_180990
|
ZACN
|
zinc activated ligand-gated ion channel
|
|
chr4_+_3738093
|
1.500
|
NM_000683
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr7_-_127458231
|
1.496
|
NM_022143
|
LRRC4
|
leucine rich repeat containing 4
|
|
chr17_+_45008175
|
1.488
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr15_+_81567327
|
1.485
|
NM_001144903
NM_023003
|
TM6SF1
|
transmembrane 6 superfamily member 1
|
|
chr22_-_37858871
|
1.484
|
|
CBX7
|
chromobox homolog 7
|
|
chr1_+_109827975
|
1.484
|
NM_153340
|
ATXN7L2
|
ataxin 7-like 2
|
|
chr8_-_142307854
|
1.484
|
NM_001080431
|
SLC45A4
|
solute carrier family 45, member 4
|
|
chr16_+_2757655
|
1.481
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr1_+_100776280
|
1.477
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr9_-_139316523
|
1.468
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr9_-_33385265
|
1.452
|
|
AQP7
|
aquaporin 7
|
|
chr2_+_241279934
|
1.441
|
NM_198998
|
AQP12A
AQP12B
|
aquaporin 12A
aquaporin 12B
|
|
chr2_-_232499202
|
1.440
|
NM_024409
|
NPPC
|
natriuretic peptide C
|
|
chr19_-_1989937
|
1.424
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr17_+_73676219
|
1.421
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr14_+_103675194
|
1.406
|
|
KIF26A
|
kinesin family member 26A
|
|
chr17_+_45993338
|
1.404
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr1_-_2451543
|
1.401
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr15_+_72906003
|
1.394
|
NM_001030005
|
CPLX3
|
complexin 3
|
|
chrX_-_16797895
|
1.391
|
NM_001198719
|
RBBP7
|
retinoblastoma binding protein 7
|
|
chr2_+_96566190
|
1.391
|
NM_212481
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like)
|
|
chr9_-_85342868
|
1.376
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr11_+_61204392
|
1.370
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr17_-_2561676
|
1.367
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr9_-_139060377
|
1.352
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr1_+_112852892
|
1.344
|
NM_024494
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr5_+_1254709
|
1.343
|
NM_001003841
|
SLC6A19
|
solute carrier family 6 (neutral amino acid transporter), member 19
|
|
chr8_-_144583718
|
1.339
|
NM_201589
|
MAFA
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian)
|
|
chr1_+_1971762
|
1.338
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr19_+_43447078
|
1.337
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr16_-_29781842
|
1.320
|
NM_006319
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase
|
|
chr20_-_538909
|
1.318
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr5_-_54317102
|
1.308
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr1_-_12599934
|
1.302
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr9_+_139006663
|
1.298
|
NM_183241
|
C9orf142
|
chromosome 9 open reading frame 142
|
|
chr17_+_45900688
|
1.295
|
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr1_+_1557419
|
1.295
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr17_-_24253821
|
1.291
|
NM_144683
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13
|
|
chr20_+_60906605
|
1.286
|
NM_007346
|
OGFR
|
opioid growth factor receptor
|
|
chr17_-_23716391
|
1.286
|
NM_001080837
|
SEBOX
|
SEBOX homeobox
|
|
chr1_-_74912009
|
1.284
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr16_+_84204424
|
1.283
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr21_+_36993860
|
1.282
|
NM_005069
NM_009586
|
SIM2
|
single-minded homolog 2 (Drosophila)
|
|
chr9_-_139060285
|
1.280
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr9_+_129893948
|
1.277
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr10_-_98935369
|
1.271
|
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr11_-_70971556
|
1.270
|
NM_001005405
|
KRTAP5-11
|
keratin associated protein 5-11
|
|
chr19_-_14177980
|
1.264
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr16_+_87765661
|
1.258
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr16_+_270606
|
1.251
|
NM_001176
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma
|
|
chrX_+_110074124
|
1.248
|
NM_001128166
NM_001128167
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr16_-_87460514
|
1.245
|
NM_001080487
|
PABPN1L
|
poly(A) binding protein, nuclear 1-like (cytoplasmic)
|
|
chr19_+_4983899
|
1.244
|
|
|
|
|
chr8_-_139995417
|
1.238
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr22_-_35011703
|
1.238
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr9_-_139042526
|
1.235
|
NM_001606
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr1_+_110456584
|
1.232
|
NM_203412
|
UBL4B
|
ubiquitin-like 4B
|
|
chr9_-_139184247
|
1.226
|
NM_001013653
|
LRRC26
|
leucine rich repeat containing 26
|
|
chr12_+_131797455
|
1.223
|
NM_001170543
NM_001170544
NM_138575
|
PGAM5
|
phosphoglycerate mutase family member 5
|
|
chr17_-_44226595
|
1.218
|
NM_173623
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr2_-_241480058
|
1.217
|
NM_024861
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr5_+_176170041
|
1.215
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr6_-_39398293
|
1.209
|
NM_001135105
NM_001135106
NM_001135107
NM_032115
|
KCNK16
|
potassium channel, subfamily K, member 16
|
|
chr13_-_49597776
|
1.208
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr9_-_130980302
|
1.206
|
|
IER5L
|
immediate early response 5-like
|
|
chr17_-_43390108
|
1.202
|
NM_024320
|
PRR15L
|
proline rich 15-like
|
|
chr8_+_143542378
|
1.199
|
NM_001702
|
BAI1
|
brain-specific angiogenesis inhibitor 1
|
|
chr19_+_12810258
|
1.196
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr17_+_24944605
|
1.193
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr19_+_54996613
|
1.191
|
|
|
|
|
chr10_-_98935570
|
1.188
|
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr11_-_64327197
|
1.183
|
NM_004579
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2
|
|
chr11_+_119616112
|
1.180
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr10_-_21847126
|
1.177
|
|
|
|
|
chr22_-_39964001
|
1.174
|
|
CHADL
|
chondroadherin-like
|
|
chr1_+_18680010
|
1.171
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr16_-_2099854
|
1.170
|
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr14_+_104261578
|
1.169
|
NM_152328
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr1_-_3556420
|
1.161
|
NM_017818
|
WDR8
|
WD repeat domain 8
|
|
chr16_-_730889
|
1.160
|
NM_022493
|
NARFL
|
nuclear prelamin A recognition factor-like
|
|
chr10_+_112247614
|
1.158
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr12_+_131705438
|
1.154
|
NM_012226
NM_016318
NM_170682
NM_170683
NM_174872
NM_174873
|
P2RX2
|
purinergic receptor P2X, ligand-gated ion channel, 2
|
|
chr2_-_96145614
|
1.152
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr19_-_50964336
|
1.148
|
NM_175875
|
SIX5
|
SIX homeobox 5
|
|
chr9_-_139042416
|
1.138
|
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr2_+_120820140
|
1.137
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr14_+_73775927
|
1.135
|
NM_182894
|
VSX2
|
visual system homeobox 2
|
|
chr5_+_132176921
|
1.135
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr11_-_64368616
|
1.132
|
NM_017525
|
CDC42BPG
|
CDC42 binding protein kinase gamma (DMPK-like)
|
|
chr3_+_50204026
|
1.126
|
NM_000172
NM_144499
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1
|
|
chr10_+_46413551
|
1.126
|
NM_014696
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2
|
|
chr9_+_101623957
|
1.126
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr19_-_60740246
|
1.123
|
NM_001101401
|
SBK2
|
SH3-binding domain kinase family, member 2
|
|
chr1_+_53300410
|
1.121
|
NM_153703
|
PODN
|
podocan
|
|
chr10_-_98470164
|
1.119
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr1_+_159943385
|
1.118
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr9_-_138757175
|
1.117
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr11_-_67163600
|
1.113
|
NM_005995
|
TBX10
|
T-box 10
|
|
chr11_+_384238
|
1.112
|
|
PKP3
|
plakophilin 3
|
|
chr17_-_77092569
|
1.111
|
|
ACTG1
|
actin, gamma 1
|
|
chr9_-_130980359
|
1.110
|
NM_203434
|
IER5L
|
immediate early response 5-like
|
|
chr11_+_63876924
|
1.107
|
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chrX_+_12066505
|
1.104
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chrX_+_109548940
|
1.103
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr19_+_13066956
|
1.102
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr19_+_18584666
|
1.096
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr11_+_56984702
|
1.092
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr13_+_26896614
|
1.085
|
NM_002097
|
GTF3A
|
general transcription factor IIIA
|
|
chr14_+_104244998
|
1.085
|
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr19_+_7834124
|
1.081
|
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr9_-_139183675
|
1.076
|
|
|
|
|
chr8_-_144882679
|
1.076
|
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr11_+_70915960
|
1.076
|
NM_001012503
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr3_+_122794859
|
1.075
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr19_+_8335010
|
1.073
|
NM_001039667
NM_139314
|
ANGPTL4
|
angiopoietin-like 4
|
|
chr8_-_29262240
|
1.072
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr19_+_10392128
|
1.068
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_-_59438411
|
1.064
|
NM_024318
|
LILRB3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr3_+_49034494
|
1.061
|
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr12_-_50871953
|
1.059
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr13_+_112681626
|
1.059
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr17_+_45008473
|
1.055
|
|
NXPH3
|
neurexophilin 3
|
|
chr2_+_10779191
|
1.055
|
NM_001039362
NM_144583
|
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2
|
|
chr17_+_4434582
|
1.054
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr17_+_21220260
|
1.053
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr11_+_384199
|
1.051
|
NM_007183
|
PKP3
|
plakophilin 3
|
|
chr5_-_148738980
|
1.050
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr5_-_525916
|
1.048
|
|
LOC25845
|
hypothetical LOC25845
|