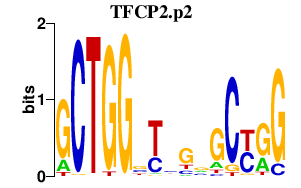
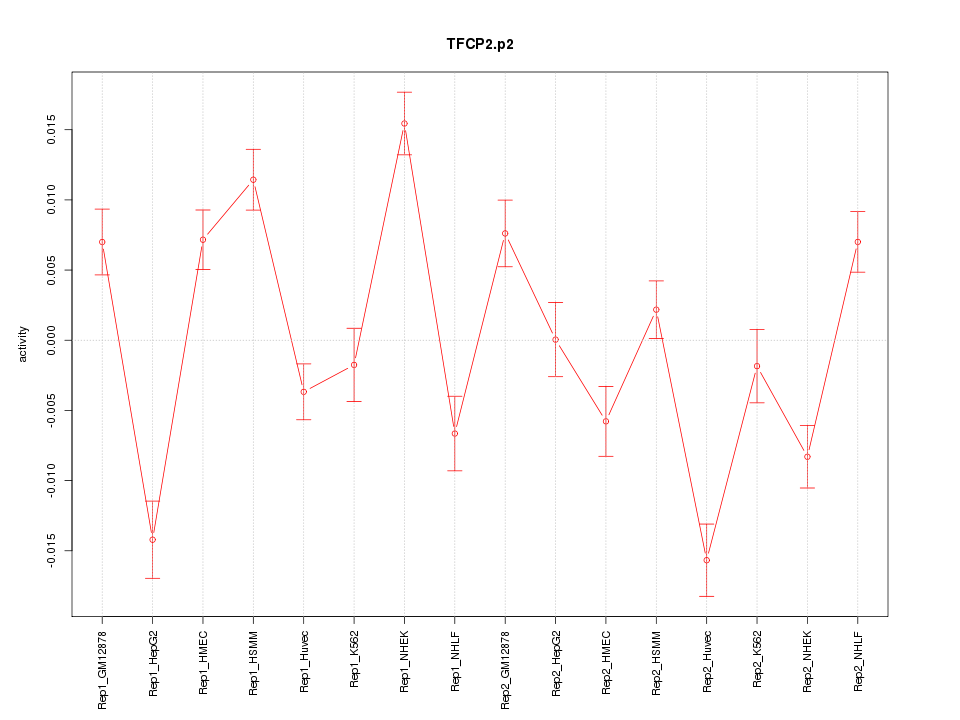
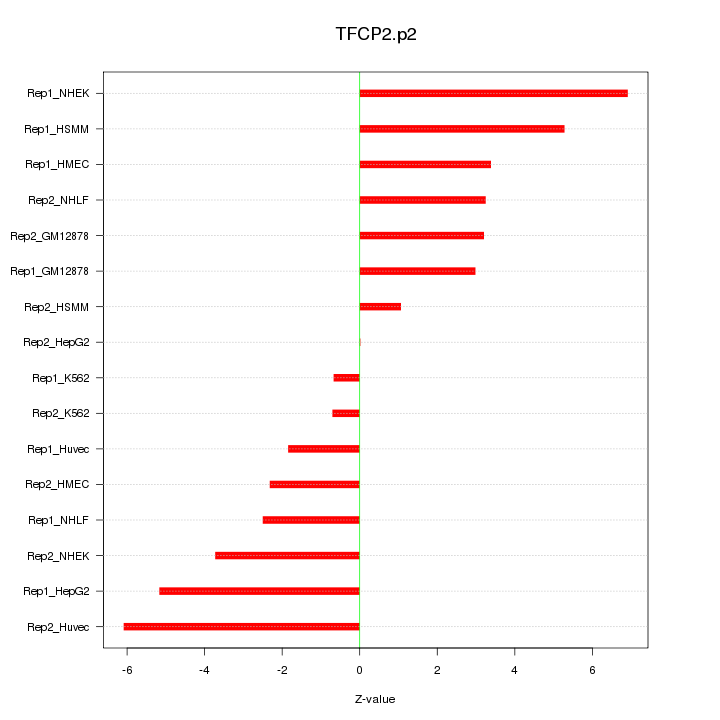
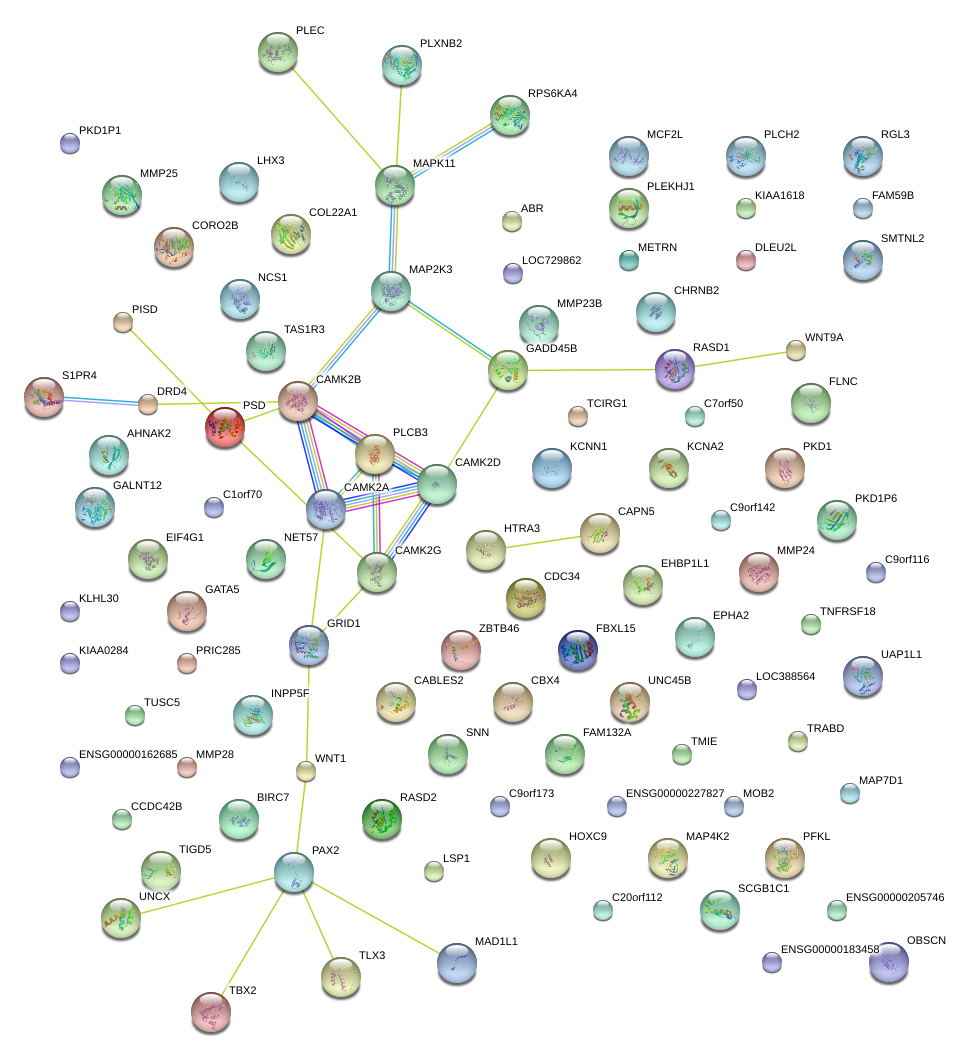
|
chr1_-_1465602
|
4.960
|
NM_001114748
|
C1orf70
|
chromosome 1 open reading frame 70
|
|
chr9_-_138234707
|
4.813
|
NM_014564
|
LHX3
|
LIM homeobox 3
|
|
chr19_-_1827133
|
3.180
|
|
|
|
|
chr11_+_65100074
|
3.134
|
NM_001099409
|
EHBP1L1
|
EH domain binding protein 1-like 1
|
|
chr20_-_30636535
|
2.985
|
|
LOC284804
|
hypothetical protein LOC284804
|
|
chr7_+_1239078
|
2.983
|
NM_001080461
|
UNCX
|
UNC homeobox
|
|
chr17_-_959007
|
2.957
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr17_-_75427644
|
2.950
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr19_-_13808101
|
2.930
|
|
LOC284454
|
hypothetical LOC284454
|
|
chr19_-_11390948
|
2.869
|
NM_001035223
NM_001161616
|
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3
|
|
chr16_-_2099854
|
2.846
|
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr12_+_47658502
|
2.831
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr13_+_112681626
|
2.704
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr11_+_627268
|
2.702
|
NM_000797
|
DRD4
|
dopamine receptor D4
|
|
chr1_+_2397613
|
2.626
|
NM_014638
|
PLCH2
|
phospholipase C, eta 2
|
|
chr17_+_1129706
|
2.619
|
NM_172367
|
TUSC5
|
tumor suppressor candidate 5
|
|
chr2_+_238712101
|
2.578
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr16_+_705080
|
2.469
|
NM_024042
|
METRN
|
meteorin, glial cell differentiation regulator
|
|
chr19_+_2427122
|
2.369
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr5_+_170668892
|
2.360
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr7_-_1946667
|
2.325
|
|
MAD1L1
|
MAD1 mitotic arrest deficient-like 1 (yeast)
|
|
chr20_+_61337679
|
2.322
|
NM_022161
NM_139317
|
BIRC7
|
baculoviral IAP repeat containing 7
|
|
chr1_-_1171964
|
2.301
|
NM_001014980
|
FAM132A
|
family with sequence similarity 132, member A
|
|
chr22_+_48971067
|
2.285
|
|
TRABD
|
TraB domain containing
|
|
chr9_+_139091759
|
2.270
|
NM_207309
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr11_+_67567022
|
2.252
|
NM_006053
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr16_+_11669736
|
2.193
|
NM_003498
|
SNN
|
stannin
|
|
chr1_-_226202210
|
2.179
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr17_+_4434582
|
2.178
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr19_+_3129735
|
2.168
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr17_+_21128614
|
2.160
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr14_-_104515738
|
2.120
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr10_-_88116181
|
2.094
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr15_+_66658626
|
2.094
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr17_+_75849221
|
2.070
|
NM_020914
NM_020954
|
RNF213
|
ring finger protein 213
|
|
chr11_+_63883192
|
2.057
|
NM_001006944
NM_003942
|
RPS6KA4
|
ribosomal protein S6 kinase, 90kDa, polypeptide 4
|
|
chr3_+_185515108
|
2.052
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr12_+_52680143
|
2.052
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr7_-_44331544
|
2.037
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr11_+_183079
|
2.027
|
NM_145651
|
SCGB1C1
|
secretoglobin, family 1C, member 1
|
|
chr1_+_36394044
|
2.025
|
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr7_+_128257698
|
2.024
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr4_+_8322358
|
2.002
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr1_+_1256585
|
1.983
|
NM_152228
|
TAS1R3
|
taste receptor, type 1, member 3
|
|
chr20_-_61669550
|
1.969
|
NM_033405
|
PRIC285
|
peroxisomal proliferator-activated receptor A interacting complex 285
|
|
chr22_-_49050847
|
1.949
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr19_-_60587414
|
1.921
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr20_-_60484420
|
1.902
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chr1_+_152806924
|
1.901
|
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr1_+_1557419
|
1.895
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr11_+_76455574
|
1.891
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr10_-_104158910
|
1.841
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr9_+_100609754
|
1.818
|
NM_024642
|
GALNT12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12)
|
|
chr12_+_112071930
|
1.772
|
NM_001144872
|
CCDC42B
|
coiled-coil domain containing 42B
|
|
chr14_+_104402626
|
1.766
|
NM_001112726
NM_015005
|
KIAA0284
|
KIAA0284
|
|
chr9_-_137531517
|
1.764
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr1_-_1131795
|
1.756
|
NM_004195
NM_148901
NM_148902
|
TNFRSF18
|
tumor necrosis factor receptor superfamily, member 18
|
|
chr1_+_152806874
|
1.752
|
NM_000748
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr17_+_21128560
|
1.738
|
NM_145109
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr8_+_144751216
|
1.737
|
NM_032862
|
TIGD5
|
tigger transposable element derived 5
|
|
chr22_+_34267297
|
1.736
|
NM_014310
|
RASD2
|
RASD family, member 2
|
|
chr2_+_26249463
|
1.732
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr10_+_102495327
|
1.730
|
NM_000278
NM_003987
NM_003988
NM_003989
NM_003990
|
PAX2
|
paired box 2
|
|
chr22_+_34266837
|
1.712
|
|
|
|
|
chr22_-_37569679
|
1.708
|
|
|
|
|
chr13_-_49597776
|
1.702
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr17_+_56832255
|
1.689
|
|
TBX2
|
T-box 2
|
|
chr20_-_60415729
|
1.683
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr17_+_30498948
|
1.680
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr11_-_64327197
|
1.668
|
NM_004579
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2
|
|
chr19_+_482688
|
1.660
|
NM_004359
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr17_+_75507750
|
1.653
|
|
|
|
|
chr17_-_17340219
|
1.642
|
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr11_+_63775567
|
1.631
|
NM_000932
NM_001184883
|
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific)
|
|
chr8_-_139995417
|
1.621
|
NM_152888
|
COL22A1
|
collagen, type XXII, alpha 1
|
|
chr1_-_110950497
|
1.616
|
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2
|
|
chr20_-_61933012
|
1.610
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr1_-_16355013
|
1.608
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr21_+_44566063
|
1.604
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr19_-_2187648
|
1.597
|
|
PLEKHJ1
|
pleckstrin homology domain containing, family J member 1
|
|
chr19_+_17923110
|
1.588
|
NM_002248
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1
|
|
chr9_+_139092016
|
1.588
|
|
UAP1L1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr22_-_49063088
|
1.577
|
|
PLXNB2
|
plexin B2
|
|
chr11_+_76491532
|
1.576
|
NM_006189
|
OMP
|
olfactory marker protein
|
|
chr3_+_46717826
|
1.571
|
NM_147196
|
TMIE
|
transmembrane inner ear
|
|
chr11_+_1848674
|
1.570
|
NM_001013254
NM_001013255
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr1_+_226462363
|
1.565
|
NM_001098623
NM_052843
|
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF
|
|
chr9_+_131974506
|
1.555
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr9_+_139265533
|
1.555
|
NM_001004353
|
C9orf173
|
chromosome 9 open reading frame 173
|
|
chr7_-_1034453
|
1.537
|
|
C7orf50
|
chromosome 7 open reading frame 50
|
|
chr16_+_3036682
|
1.535
|
NM_022468
|
MMP25
|
matrix metallopeptidase 25
|
|
chr9_+_139006663
|
1.534
|
NM_183241
|
C9orf142
|
chromosome 9 open reading frame 142
|
|
chr8_-_145099963
|
1.532
|
NM_201379
|
PLEC
|
plectin
|
|
chr17_+_21128625
|
1.526
|
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr11_-_1464386
|
1.523
|
NM_001172223
|
MOB2
|
Mps one binder kinase activator-like 2
|
|
chr21_+_46226072
|
1.521
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr10_-_125641484
|
1.519
|
NM_198148
|
CPXM2
|
carboxypeptidase X (M14 family), member 2
|
|
chr20_-_26137755
|
1.513
|
|
LOC284801
|
hypothetical protein LOC284801
|
|
chrX_+_150614385
|
1.508
|
NM_024082
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
|
|
chr7_-_100331417
|
1.505
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr1_+_226741665
|
1.493
|
NM_001010858
|
RNF187
|
ring finger protein 187
|
|
chr15_+_29406335
|
1.489
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr9_+_36126710
|
1.486
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr12_-_51585004
|
1.483
|
NM_002273
|
KRT8
|
keratin 8
|
|
chr9_-_95757448
|
1.472
|
|
BARX1
|
BARX homeobox 1
|
|
chr2_+_219991342
|
1.466
|
NM_001927
|
DES
|
desmin
|
|
chr17_-_45562112
|
1.463
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr16_+_1949517
|
1.463
|
NM_004548
|
NDUFB10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa
|
|
chr11_+_65802221
|
1.461
|
NM_182553
|
CNIH2
|
cornichon homolog 2 (Drosophila)
|
|
chr19_+_2187815
|
1.452
|
NM_007165
|
SF3A2
|
splicing factor 3a, subunit 2, 66kDa
|
|
chr9_-_139184247
|
1.443
|
NM_001013653
|
LRRC26
|
leucine rich repeat containing 26
|
|
chr17_+_24095135
|
1.440
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr11_+_64838882
|
1.414
|
NM_006779
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chr2_+_241023908
|
1.413
|
|
GPC1
|
glypican 1
|
|
chr19_+_482737
|
1.408
|
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chrX_+_153339814
|
1.407
|
NM_017514
|
PLXNA3
|
plexin A3
|
|
chr19_+_54872304
|
1.402
|
NM_001536
NM_198318
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr19_+_40824486
|
1.402
|
NM_014209
|
ETV2
|
ets variant 2
|
|
chr10_+_124210980
|
1.398
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr11_-_2136129
|
1.391
|
|
|
|
|
chr22_-_37569962
|
1.391
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr11_-_62539857
|
1.389
|
NM_001184732
NM_001184733
NM_001184736
NM_004254
|
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8
|
|
chr9_+_36126667
|
1.387
|
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr15_-_26017994
|
1.370
|
NM_000275
|
OCA2
|
oculocutaneous albinism II
|
|
chr14_-_99842519
|
1.370
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr16_+_670420
|
1.369
|
|
STUB1
|
STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase
|
|
chr9_-_95757365
|
1.366
|
NM_021570
|
BARX1
|
BARX homeobox 1
|
|
chr19_+_54872220
|
1.362
|
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr11_-_407324
|
1.361
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr19_-_50429306
|
1.361
|
NM_138568
|
EXOC3L2
|
exocyst complex component 3-like 2
|
|
chr8_+_145718702
|
1.356
|
|
|
|
|
chr9_+_125813849
|
1.352
|
|
LHX2
|
LIM homeobox 2
|
|
chr17_+_56831951
|
1.348
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr1_+_154478574
|
1.345
|
NM_199173
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein
|
|
chr9_+_137107007
|
1.342
|
|
OLFM1
|
olfactomedin 1
|
|
chr19_+_590878
|
1.339
|
NM_020637
|
FGF22
|
fibroblast growth factor 22
|
|
chr16_-_2521359
|
1.336
|
NM_001048212
|
CEMP1
|
cementum protein 1
|
|
chr2_+_232951487
|
1.333
|
NM_001632
|
ALPP
|
alkaline phosphatase, placental
|
|
chr2_+_71016503
|
1.323
|
NM_001692
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1
|
|
chr22_-_39964001
|
1.320
|
|
CHADL
|
chondroadherin-like
|
|
chr9_+_137107187
|
1.317
|
|
OLFM1
|
olfactomedin 1
|
|
chr9_+_137106909
|
1.316
|
NM_014279
NM_006334
|
OLFM1
|
olfactomedin 1
|
|
chr16_+_670426
|
1.313
|
|
STUB1
|
STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase
|
|
chr4_+_4912272
|
1.308
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr2_-_128792515
|
1.305
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr16_+_2226468
|
1.296
|
NM_001374
|
DNASE1L2
|
deoxyribonuclease I-like 2
|
|
chr9_+_91409746
|
1.290
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr17_+_72982919
|
1.288
|
NM_001113495
|
SEPT9
|
septin 9
|
|
chr19_-_63550712
|
1.287
|
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr1_+_1099145
|
1.280
|
NM_001130045
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr2_+_241023703
|
1.279
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr8_+_145705253
|
1.277
|
NM_138431
|
MFSD3
|
major facilitator superfamily domain containing 3
|
|
chr16_+_670389
|
1.273
|
|
STUB1
|
STIP1 homology and U-box containing protein 1, E3 ubiquitin protein ligase
|
|
chr9_+_137107088
|
1.273
|
|
OLFM1
|
olfactomedin 1
|
|
chr1_-_25129354
|
1.267
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr10_+_133850325
|
1.266
|
NM_006426
|
DPYSL4
|
dihydropyrimidinase-like 4
|
|
chr1_+_36394338
|
1.266
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr1_-_25129305
|
1.266
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr6_+_150326802
|
1.264
|
NM_025218
|
ULBP1
|
UL16 binding protein 1
|
|
chr11_-_785005
|
1.257
|
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22
|
|
chr19_+_60816770
|
1.250
|
NM_001195605
|
ZNF865
|
zinc finger protein 865
|
|
chrX_+_128942435
|
1.242
|
|
|
|
|
chr20_-_62151216
|
1.241
|
NM_018419
|
SOX18
|
SRY (sex determining region Y)-box 18
|
|
chr10_+_112247614
|
1.241
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr22_-_22252376
|
1.240
|
NM_020070
NM_152855
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1
|
|
chr19_-_19615369
|
1.240
|
|
GMIP
|
GEM interacting protein
|
|
chr17_+_56832492
|
1.237
|
|
TBX2
|
T-box 2
|
|
chr14_+_102659464
|
1.233
|
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr10_+_104145421
|
1.230
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr20_+_55399847
|
1.225
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr1_+_200124412
|
1.223
|
NM_198149
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr10_-_99521701
|
1.222
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr17_+_7225088
|
1.222
|
NM_003985
|
TNK1
|
tyrosine kinase, non-receptor, 1
|
|
chr19_+_1397268
|
1.220
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr1_+_39229474
|
1.208
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr17_+_38250122
|
1.204
|
NM_001158
NM_009590
|
AOC2
|
amine oxidase, copper containing 2 (retina-specific)
|
|
chr1_+_968522
|
1.203
|
|
AGRN
|
agrin
|
|
chr16_+_1323627
|
1.203
|
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr19_-_19615452
|
1.202
|
NM_016573
|
GMIP
|
GEM interacting protein
|
|
chr16_+_2593351
|
1.196
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr11_+_46339691
|
1.196
|
NM_001105540
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr16_-_87646856
|
1.194
|
|
|
|
|
chr2_-_27339278
|
1.193
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr8_-_144969529
|
1.192
|
NM_015356
NM_182706
|
SCRIB
|
scribbled homolog (Drosophila)
|
|
chr10_+_134901397
|
1.187
|
NM_014468
|
VENTX
|
VENT homeobox homolog (Xenopus laevis)
|
|
chr1_+_39729885
|
1.185
|
NM_181809
|
BMP8A
|
bone morphogenetic protein 8a
|
|
chr11_+_35117408
|
1.173
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr11_+_2355099
|
1.173
|
|
CD81
|
CD81 molecule
|
|
chr17_-_42250961
|
1.168
|
NM_030753
|
WNT3
|
wingless-type MMTV integration site family, member 3
|
|
chr17_-_60208083
|
1.167
|
|
LOC146880
|
hypothetical LOC146880
|
|
chr19_-_7896974
|
1.162
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr16_-_88314762
|
1.161
|
NM_004913
|
C16orf7
|
chromosome 16 open reading frame 7
|
|
chr16_+_82559600
|
1.159
|
NM_019065
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr4_+_7096043
|
1.159
|
NM_152293
|
TADA2B
|
transcriptional adaptor 2B
|
|
chr10_+_6284845
|
1.156
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr9_+_137107222
|
1.154
|
|
OLFM1
|
olfactomedin 1
|
|
chr9_-_35062643
|
1.149
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr20_+_60311443
|
1.148
|
|
ADRM1
|
adhesion regulating molecule 1
|
|
chr11_+_35117391
|
1.148
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr20_+_60311334
|
1.148
|
|
ADRM1
|
adhesion regulating molecule 1
|