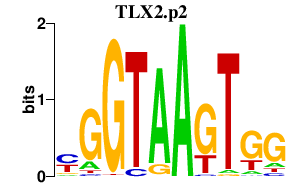
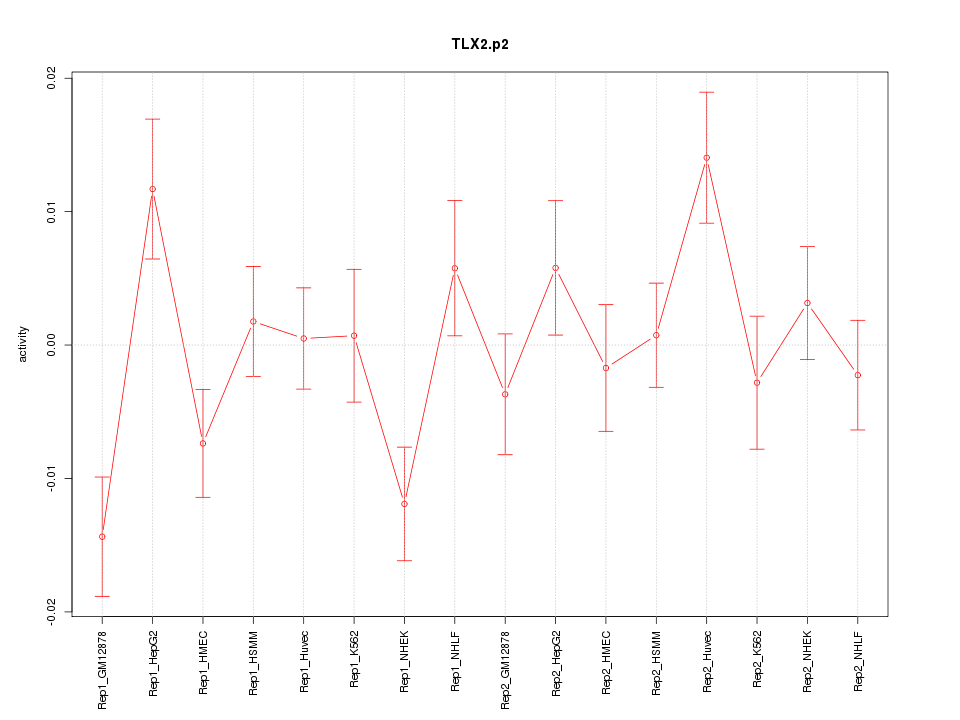
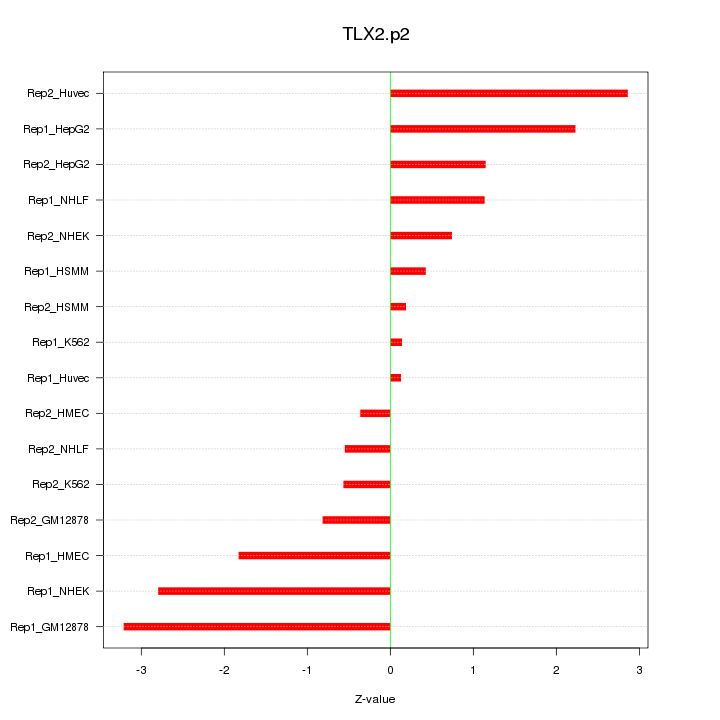
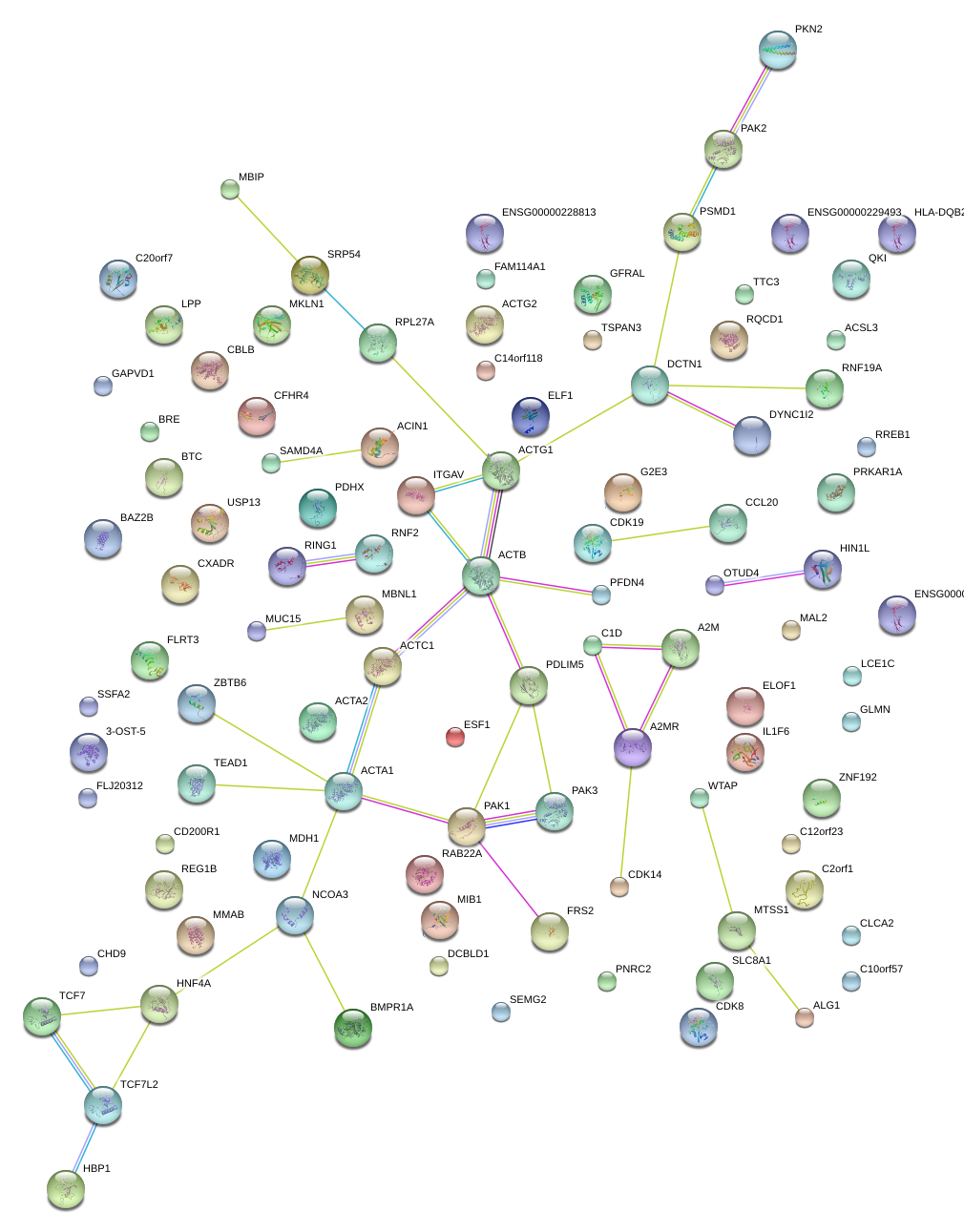
|
chr14_+_54104383
|
2.244
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr3_+_153469421
|
1.506
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr4_+_95592067
|
1.470
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr12_-_9123126
|
1.400
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr7_+_106596641
|
1.382
|
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr14_+_34521945
|
1.253
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr4_+_95592030
|
1.161
|
NM_001011513
NM_001011515
NM_001011516
NM_006457
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr12_+_68150332
|
1.138
|
NM_001042555
NM_006654
|
FRS2
|
fibroblast growth factor receptor substrate 2
|
|
chr2_+_172252194
|
1.107
|
NM_001378
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2
|
|
chr10_-_90702490
|
1.099
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr14_+_54104099
|
1.091
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr14_+_34521854
|
1.081
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr2_-_63669265
|
1.074
|
NM_015910
|
WDPCP
|
WD repeat containing planar cell polarity effector
|
|
chr20_-_14266200
|
1.074
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr2_+_187163232
|
1.047
|
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr12_+_105873669
|
1.040
|
NM_152261
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr13_+_25726755
|
1.040
|
NM_001260
|
CDK8
|
cyclin-dependent kinase 8
|
|
chr3_-_153470022
|
1.035
|
|
LOC401093
|
hypothetical LOC401093
|
|
chr2_+_63669596
|
1.025
|
NM_005917
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr1_+_183281173
|
1.016
|
NM_007212
|
RNF2
|
ring finger protein 2
|
|
chr11_+_34894688
|
1.007
|
|
PDHX
|
pyruvate dehydrogenase complex, component X
|
|
chr20_+_43283423
|
0.979
|
NM_003008
|
SEMG2
|
semenogelin II
|
|
chr4_-_75938710
|
0.960
|
NM_001729
|
BTC
|
betacellulin
|
|
chr21_+_17841297
|
0.941
|
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr14_-_35859545
|
0.935
|
NM_001144891
NM_016586
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr2_-_160277083
|
0.922
|
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr2_-_74460933
|
0.912
|
NM_001135040
NM_001190837
NM_004082
|
DCTN1
|
dynactin 1
|
|
chr16_+_51722285
|
0.912
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr1_-_92537087
|
0.908
|
NM_053274
|
GLMN
|
glomulin, FKBP associated protein
|
|
chr4_+_38545741
|
0.889
|
NM_138389
|
FAM114A1
|
family with sequence similarity 114, member A1
|
|
chr2_+_207215386
|
0.887
|
NM_001102659
|
LOC200726
|
hCG1657980
|
|
chr2_+_228386800
|
0.886
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr6_+_117910472
|
0.875
|
NM_173674
|
DCBLD1
|
discoidin, CUB and LCCL domain containing 1
|
|
chr17_+_64020120
|
0.860
|
NM_002734
NM_212472
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr2_+_223433919
|
0.848
|
NM_004457
NM_203372
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3
|
|
chr13_+_96797290
|
0.847
|
|
|
|
|
chr20_+_56318415
|
0.837
|
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr14_+_30098079
|
0.836
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr13_-_40491491
|
0.834
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr11_+_12652427
|
0.827
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr6_+_131190237
|
0.826
|
NM_001195597
|
LOC285733
|
hypothetical LOC285733
|
|
chr2_+_187162969
|
0.826
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr20_+_52258023
|
0.816
|
|
PFDN4
|
prefoldin subunit 4
|
|
chr8_-_125785611
|
0.810
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr20_-_13713509
|
0.810
|
NM_016649
|
ESF1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae)
|
|
chr1_-_151045730
|
0.808
|
NM_178351
|
LCE1C
|
late cornified envelope 1C
|
|
chr20_+_45564063
|
0.805
|
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr2_+_219141761
|
0.803
|
NM_005444
|
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe)
|
|
chr6_+_28217590
|
0.802
|
NM_006298
|
ZNF192
|
zinc finger protein 192
|
|
chr17_+_64020155
|
0.797
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr6_+_160068135
|
0.796
|
NM_004906
NM_152858
|
WTAP
|
Wilms tumor 1 associated protein
|
|
chr11_+_8660913
|
0.789
|
|
RPL27A
|
ribosomal protein L27a
|
|
chr3_+_189425886
|
0.788
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr2_-_79168626
|
0.786
|
NM_006507
|
REG1B
|
regenerating islet-derived 1 beta
|
|
chr2_+_231629839
|
0.782
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr18_+_17575542
|
0.782
|
NM_020774
|
MIB1
|
mindbomb homolog 1 (Drosophila)
|
|
chr2_+_113479919
|
0.779
|
NM_014440
|
IL1F6
|
interleukin 1 family, member 6 (epsilon)
|
|
chr1_+_24158898
|
0.778
|
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr6_+_163904564
|
0.763
|
|
QKI
|
quaking homolog, KH domain RNA binding (mouse)
|
|
chr8_+_120289735
|
0.762
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr3_-_114176401
|
0.760
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr1_+_86662356
|
0.757
|
NM_006536
|
CLCA2
|
chloride channel accessory 2
|
|
chr2_-_40593074
|
0.756
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr2_+_231629806
|
0.755
|
NM_001191037
NM_002807
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr14_-_22610610
|
0.754
|
NM_001164816
NM_001164817
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr3_-_107070400
|
0.744
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr10_+_82163568
|
0.743
|
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr20_+_13713645
|
0.741
|
NM_001039375
NM_024120
|
C20orf7
|
chromosome 20 open reading frame 7
|
|
chr17_+_64020159
|
0.737
|
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr6_-_32839244
|
0.732
|
NM_001198858
|
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2
|
|
chr12_+_55808696
|
0.726
|
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr20_+_45564007
|
0.724
|
NM_001174087
NM_001174088
NM_006534
NM_181659
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr13_-_40491405
|
0.715
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr20_+_42463279
|
0.712
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr17_+_55269974
|
0.711
|
|
|
|
|
chr16_+_5061862
|
0.711
|
|
ALG1
|
asparagine-linked glycosylation 1, beta-1,4-mannosyltransferase homolog (S. cerevisiae)
|
|
chr6_+_55300225
|
0.707
|
NM_207410
|
GFRAL
|
GDNF family receptor alpha like
|
|
chr15_-_75135496
|
0.706
|
|
TSPAN3
|
tetraspanin 3
|
|
chr20_+_42463419
|
0.704
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr21_+_37367432
|
0.702
|
NM_001001894
|
TTC3
|
tetratricopeptide repeat domain 3
|
|
chr1_+_195123766
|
0.698
|
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr3_+_180853473
|
0.698
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr14_+_75688042
|
0.693
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr9_-_124715392
|
0.691
|
NM_006626
|
ZBTB6
|
zinc finger and BTB domain containing 6
|
|
chr7_+_90063543
|
0.690
|
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr11_-_26550243
|
0.688
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr4_-_146315341
|
0.688
|
NM_017493
|
OTUD4
|
OTU domain containing 4
|
|
chr14_-_22610572
|
0.682
|
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr1_+_24158887
|
0.681
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr3_+_197951203
|
0.679
|
|
PAK2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr7_+_130663161
|
0.677
|
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr10_+_88506320
|
0.676
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr6_-_58395610
|
0.676
|
|
GUSBP4
|
glucuronidase, beta pseudogene 4
|
|
chr10_+_114700775
|
0.672
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr8_-_101384633
|
0.672
|
NM_015435
|
RNF19A
|
ring finger protein 19A
|
|
chr2_+_27848085
|
0.667
|
NM_004891
NM_145330
|
MRPL33
|
mitochondrial ribosomal protein L33
|
|
chr2_+_27848090
|
0.666
|
|
MRPL33
|
mitochondrial ribosomal protein L33
|
|
chr20_+_42463350
|
0.666
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr6_+_7052798
|
0.664
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr2_+_182464910
|
0.663
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr14_+_75687997
|
0.656
|
NM_017926
NM_017972
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr2_+_32244406
|
0.651
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr1_+_114273961
|
0.646
|
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr19_-_56726704
|
0.642
|
NM_001177547
NM_001177548
NM_001177549
NM_001245
NM_198845
NM_198846
|
SIGLEC6
|
sialic acid binding Ig-like lectin 6
|
|
chr3_+_87359088
|
0.640
|
NM_014043
|
CHMP2B
|
chromatin modifying protein 2B
|
|
chr20_+_42463387
|
0.639
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr14_+_75688025
|
0.638
|
|
C14orf118
|
chromosome 14 open reading frame 118
|
|
chr5_+_146594718
|
0.636
|
NM_001112724
NM_145001
|
STK32A
|
serine/threonine kinase 32A
|
|
chr6_+_124166767
|
0.636
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr20_+_43269051
|
0.634
|
NM_003007
|
SEMG1
|
semenogelin I
|
|
chr1_+_208178160
|
0.631
|
NM_001146261
NM_001146262
NM_001146264
NM_153262
|
SYT14
|
synaptotagmin XIV
|
|
chr20_+_52257800
|
0.631
|
NM_002623
|
PFDN4
|
prefoldin subunit 4
|
|
chr6_+_117109042
|
0.624
|
NM_002269
|
KPNA5
|
karyopherin alpha 5 (importin alpha 6)
|
|
chr10_+_123862430
|
0.623
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr15_+_46285789
|
0.622
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr20_+_42947764
|
0.622
|
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr16_+_55963899
|
0.621
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr7_-_80386258
|
0.619
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr7_+_130663134
|
0.616
|
NM_013255
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr1_-_94779727
|
0.615
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr3_+_187813538
|
0.612
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr3_+_187813553
|
0.611
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr14_-_24148658
|
0.610
|
NM_033423
|
GZMH
|
granzyme H (cathepsin G-like 2, protein h-CCPX)
|
|
chr17_+_19853200
|
0.609
|
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr4_+_146250978
|
0.608
|
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr9_+_132579088
|
0.607
|
NM_007313
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase
|
|
chr19_+_2818332
|
0.606
|
NM_024967
|
ZNF556
|
zinc finger protein 556
|
|
chr22_+_29974368
|
0.605
|
|
LIMK2
|
LIM domain kinase 2
|
|
chr3_-_15814678
|
0.604
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr3_+_12305336
|
0.602
|
NM_138711
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr14_+_38804226
|
0.597
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr13_+_40783340
|
0.595
|
NM_001110798
NM_024561
NM_018527
|
NAA16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit
|
|
chr4_+_38722593
|
0.594
|
NM_001007075
NM_001171654
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr4_+_123311207
|
0.593
|
NM_015312
|
KIAA1109
|
KIAA1109
|
|
chr2_+_231629869
|
0.589
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr16_+_55963883
|
0.588
|
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr20_-_61028263
|
0.587
|
|
DIDO1
|
death inducer-obliterator 1
|
|
chr5_+_149845503
|
0.587
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr18_+_17575275
|
0.586
|
|
MIB1
|
mindbomb homolog 1 (Drosophila)
|
|
chr21_+_29592984
|
0.585
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr14_+_88099443
|
0.585
|
NM_207661
|
ZC3H14
|
zinc finger CCCH-type containing 14
|
|
chr12_-_48508411
|
0.585
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chrX_+_154097894
|
0.581
|
NM_003372
|
VBP1
|
von Hippel-Lindau binding protein 1
|
|
chr6_+_147568795
|
0.578
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr21_+_29593088
|
0.574
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr20_-_61028287
|
0.573
|
|
DIDO1
|
death inducer-obliterator 1
|
|
chr2_+_160277219
|
0.571
|
NM_022826
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7
|
|
chr1_+_84316217
|
0.570
|
NM_002731
NM_207578
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_+_187813405
|
0.569
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr3_+_58452560
|
0.562
|
NM_001128214
|
KCTD6
|
potassium channel tetramerisation domain containing 6
|
|
chr15_+_38296920
|
0.560
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr11_-_22807842
|
0.560
|
NM_148893
|
SVIP
|
small VCP/p97-interacting protein
|
|
chr3_+_150330060
|
0.556
|
NM_032383
|
HPS3
|
Hermansky-Pudlak syndrome 3
|
|
chr21_+_29593607
|
0.556
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr2_-_210798232
|
0.555
|
NM_001608
|
ACADL
|
acyl-CoA dehydrogenase, long chain
|
|
chr14_+_64041170
|
0.554
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr14_+_64041257
|
0.552
|
|
ZBTB1
|
zinc finger and BTB domain containing 1
|
|
chr2_+_231629852
|
0.552
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr6_-_112186977
|
0.550
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr12_+_79995927
|
0.550
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr20_-_61028319
|
0.549
|
NM_001193369
NM_001193370
NM_080796
|
DIDO1
|
death inducer-obliterator 1
|
|
chr18_+_3237815
|
0.549
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr2_+_160277273
|
0.549
|
|
MARCH7
|
membrane-associated ring finger (C3HC4) 7
|
|
chr12_+_67426225
|
0.546
|
|
SLC35E3
|
solute carrier family 35, member E3
|
|
chr3_-_21767819
|
0.544
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr13_+_59869427
|
0.542
|
NM_001146070
|
TDRD3
|
tudor domain containing 3
|
|
chr18_+_3237774
|
0.542
|
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric
|
|
chr12_+_84198015
|
0.542
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr14_-_22825148
|
0.541
|
NM_020834
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr6_+_133177399
|
0.537
|
NM_001016
|
SNORA33
RPS12
|
small nucleolar RNA, H/ACA box 33
ribosomal protein S12
|
|
chr21_+_29593098
|
0.536
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr3_+_153469206
|
0.535
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr1_+_208178198
|
0.535
|
|
SYT14
|
synaptotagmin XIV
|
|
chr7_+_30034731
|
0.534
|
|
PLEKHA8P1
PLEKHA8
|
pleckstrin homology domain containing, family A member 8 pseudogene 1
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8
|
|
chr6_+_64403936
|
0.532
|
|
PHF3
|
PHD finger protein 3
|
|
chr18_+_27332024
|
0.531
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr16_-_66558197
|
0.525
|
NM_001145964
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr16_-_57309625
|
0.525
|
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chr8_-_82186832
|
0.524
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr5_-_13997588
|
0.522
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr6_+_7053183
|
0.522
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr3_+_101462375
|
0.522
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr2_+_60962279
|
0.521
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr3_+_140545528
|
0.520
|
NM_020191
|
MRPS22
|
mitochondrial ribosomal protein S22
|
|
chr14_-_22825072
|
0.518
|
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr10_+_31648103
|
0.517
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr3_+_4996141
|
0.516
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr20_+_47328585
|
0.513
|
|
NCRNA00275
|
non-protein coding RNA 275
|
|
chr8_+_26491507
|
0.512
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr5_+_118434966
|
0.510
|
NM_005509
|
DMXL1
|
Dmx-like 1
|
|
chr3_+_4996305
|
0.509
|
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr13_+_26029879
|
0.508
|
|
WASF3
|
WAS protein family, member 3
|
|
chr4_+_86968069
|
0.508
|
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr20_+_47328575
|
0.504
|
|
NCRNA00275
|
non-protein coding RNA 275
|
|
chr20_+_42947754
|
0.503
|
NM_003404
NM_139323
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide
|
|
chr20_-_1420168
|
0.502
|
NM_001122962
NM_001134836
|
SIRPB2
|
signal-regulatory protein beta 2
|
|
chr8_+_97575370
|
0.499
|
|
SDC2
|
syndecan 2
|
|
chr4_+_25931725
|
0.495
|
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region
|
|
chr14_+_23008742
|
0.495
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr2_+_152900076
|
0.495
|
|
FMNL2
|
formin-like 2
|