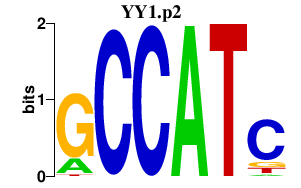
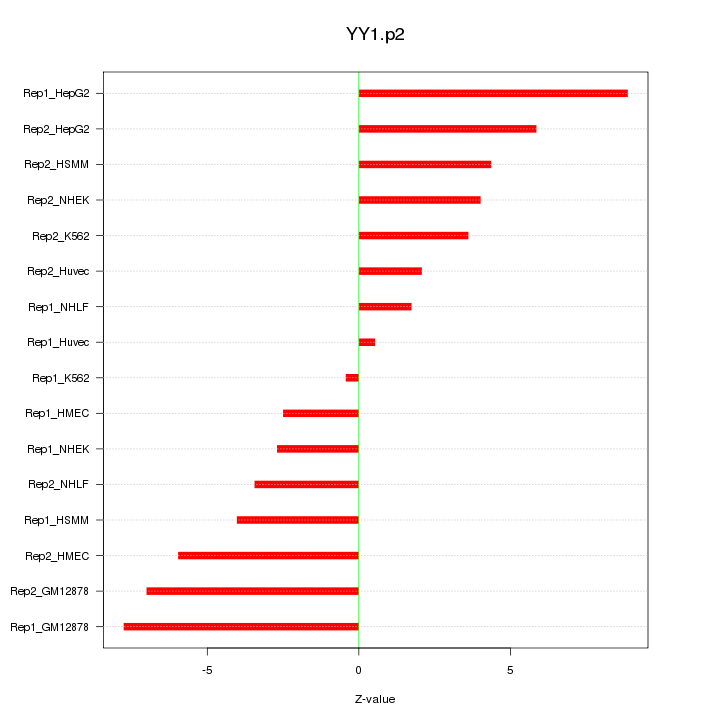
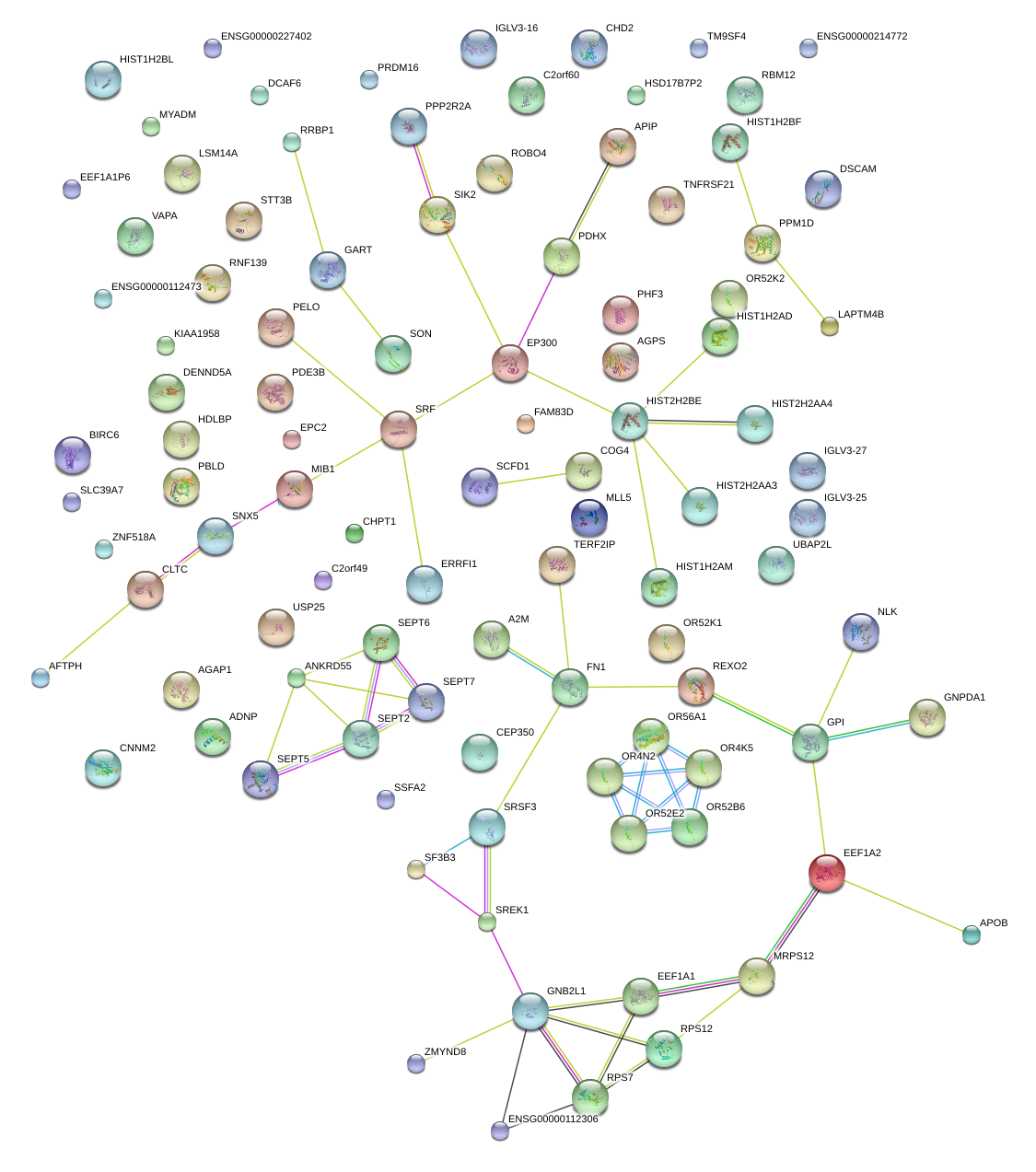
|
chr6_-_74284522
|
5.649
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr1_-_7998265
|
5.558
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr20_-_33706532
|
4.516
|
|
RBM12
|
RNA binding motif protein 12
|
|
chr12_+_100615530
|
4.444
|
NM_020244
|
CHPT1
|
choline phosphotransferase 1
|
|
chr17_+_55052185
|
3.966
|
|
|
|
|
chr8_+_26204921
|
3.903
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr3_+_31549478
|
3.821
|
NM_178862
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr2_-_215949566
|
3.740
|
|
FN1
|
fibronectin 1
|
|
chr3_+_31549435
|
3.724
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr8_+_26204997
|
3.711
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr1_-_148080888
|
3.668
|
NM_001040874
NM_003516
|
HIST2H2AA4
HIST2H2AA3
|
histone cluster 2, H2aa4
histone cluster 2, H2aa3
|
|
chr2_+_149119029
|
3.634
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr17_+_55052065
|
3.602
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr19_+_59068787
|
3.552
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr8_+_26204960
|
3.528
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr10_-_69762654
|
3.503
|
NM_001033083
NM_022129
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr16_+_74239135
|
3.446
|
NM_018975
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein
|
|
chr17_+_23394062
|
3.424
|
|
NLK
|
nemo-like kinase
|
|
chr6_-_47362102
|
3.376
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr2_-_21106291
|
3.256
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr2_+_149118791
|
3.171
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr8_+_125556374
|
3.122
|
|
RNF139
|
ring finger protein 139
|
|
chr10_+_97879457
|
3.111
|
NM_014803
|
ZNF518A
|
zinc finger protein 518A
|
|
chr11_-_5037432
|
3.097
|
NM_001005164
|
OR52E2
|
olfactory receptor, family 52, subfamily E, member 2
|
|
chr9_+_114376709
|
3.080
|
|
KIAA1958
|
KIAA1958
|
|
chr1_+_178190493
|
3.045
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr20_-_17589122
|
3.039
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr20_-_17897067
|
2.998
|
NM_014426
|
SNX5
|
sorting nexin 5
|
|
chr16_+_69115227
|
2.971
|
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr20_+_30160952
|
2.966
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr3_+_31549123
|
2.949
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr7_+_104441730
|
2.942
|
NM_018682
NM_182931
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr6_-_47361921
|
2.936
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr2_-_21081694
|
2.929
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr5_-_180603335
|
2.897
|
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1
|
|
chr17_+_23393744
|
2.878
|
NM_016231
|
NLK
|
nemo-like kinase
|
|
chr20_-_17897384
|
2.839
|
NM_152227
|
SNX5
|
sorting nexin 5
|
|
chr8_+_125556294
|
2.824
|
|
RNF139
|
ring finger protein 139
|
|
chr2_-_21087435
|
2.788
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr5_+_52131686
|
2.772
|
|
PELO
|
pelota homolog (Drosophila)
|
|
chr21_+_16024157
|
2.757
|
|
USP25
|
ubiquitin specific peptidase 25
|
|
chr15_+_91244422
|
2.749
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr2_+_241903943
|
2.730
|
NM_001008492
NM_004404
|
SEPT2
|
septin 2
|
|
chr17_+_55052060
|
2.713
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr2_+_3600984
|
2.676
|
|
RPS7
|
ribosomal protein S7
|
|
chr10_+_97879645
|
2.669
|
|
ZNF518A
|
zinc finger protein 518A
|
|
chr17_+_55052046
|
2.662
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr2_+_182464910
|
2.641
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr15_+_91244563
|
2.627
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr11_+_113815317
|
2.606
|
NM_015523
|
REXO2
|
REX2, RNA exonuclease 2 homolog (S. cerevisiae)
|
|
chr11_+_4466684
|
2.579
|
NM_001005171
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1
|
|
chr2_+_3600784
|
2.575
|
|
RPS7
|
ribosomal protein S7
|
|
chr6_+_33276580
|
2.544
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr11_+_34894688
|
2.539
|
|
PDHX
|
pyruvate dehydrogenase complex, component X
|
|
chr20_+_30161173
|
2.535
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr11_-_6005546
|
2.525
|
NM_001001917
|
OR56A1
|
olfactory receptor, family 56, subfamily A, member 1
|
|
chr2_+_64605049
|
2.522
|
|
AFTPH
|
aftiphilin
|
|
chr21_+_33837233
|
2.512
|
|
SON
|
SON DNA binding protein
|
|
chr2_+_3600721
|
2.509
|
NM_001011
|
RPS7
|
ribosomal protein S7
|
|
chr8_+_98857190
|
2.502
|
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr8_+_125556130
|
2.473
|
NM_007218
|
RNF139
|
ring finger protein 139
|
|
chr5_-_180603476
|
2.469
|
NM_006098
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1
|
|
chr2_-_177965626
|
2.450
|
|
LOC100130691
|
hypothetical LOC100130691
|
|
chr2_+_177965617
|
2.424
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr2_+_3600765
|
2.419
|
|
RPS7
|
ribosomal protein S7
|
|
chr5_+_65476418
|
2.411
|
NM_139168
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr21_+_33837215
|
2.405
|
NM_032195
NM_138927
|
SON
|
SON DNA binding protein
|
|
chr11_+_5558682
|
2.395
|
NM_001005162
|
OR52B6
|
olfactory receptor, family 52, subfamily B, member 6
|
|
chr2_+_32435800
|
2.375
|
|
BIRC6
|
baculoviral IAP repeat containing 6
|
|
chr19_+_39355254
|
2.368
|
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr21_+_33837212
|
2.362
|
|
SON
|
SON DNA binding protein
|
|
chr1_+_152459278
|
2.359
|
NM_001127320
|
UBAP2L
|
ubiquitin associated protein 2-like
|
|
chr21_-_33836994
|
2.352
|
NM_001136005
NM_001136006
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
|
|
chr2_+_32435550
|
2.332
|
NM_016252
|
BIRC6
|
baculoviral IAP repeat containing 6
|
|
chr2_+_105320440
|
2.320
|
NM_024093
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr14_+_19365447
|
2.316
|
NM_001004723
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2
|
|
chr16_+_69115189
|
2.294
|
NM_012426
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr2_+_64604940
|
2.292
|
NM_001002243
NM_017657
NM_203437
|
AFTPH
|
aftiphilin
|
|
chr17_+_56032385
|
2.290
|
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr14_+_30161285
|
2.273
|
|
SCFD1
|
sec1 family domain containing 1
|
|
chr16_+_69115225
|
2.249
|
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr2_+_105320247
|
2.239
|
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr16_-_69114931
|
2.238
|
NM_001195139
NM_015386
|
COG4
|
component of oligomeric golgi complex 4
|
|
chr10_+_104667991
|
2.232
|
NM_017649
NM_199076
NM_199077
|
CNNM2
|
cyclin M2
|
|
chr19_+_39355151
|
2.224
|
NM_001114093
NM_015578
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr19_+_39355280
|
2.219
|
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr11_-_34894407
|
2.211
|
NM_015957
|
APIP
|
APAF1 interacting protein
|
|
chr14_+_30161267
|
2.193
|
NM_016106
NM_182835
|
SCFD1
|
sec1 family domain containing 1
|
|
chr6_+_64403685
|
2.186
|
|
PHF3
|
PHD finger protein 3
|
|
chr22_+_21359175
|
2.153
|
|
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr12_-_9123126
|
2.146
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr6_+_133177403
|
2.140
|
|
RPS12
|
ribosomal protein S12
|
|
chr20_-_48980858
|
2.113
|
NM_015339
NM_181442
|
ADNP
|
activity-dependent neuroprotector homeobox
|
|
chr6_+_64403631
|
2.112
|
|
PHF3
|
PHD finger protein 3
|
|
chr6_-_27883662
|
2.106
|
NM_003519
|
HIST1H2BL
|
histone cluster 1, H2bl
|
|
chr2_+_177965734
|
2.105
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr11_+_14621972
|
2.104
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr6_+_133177399
|
2.103
|
NM_001016
|
SNORA33
RPS12
|
small nucleolar RNA, H/ACA box 33
ribosomal protein S12
|
|
chr22_+_39818553
|
2.101
|
NM_001429
|
EP300
|
E1A binding protein p300
|
|
chr2_-_241903925
|
2.065
|
NM_005336
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr17_+_55051818
|
2.047
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr6_+_36670113
|
2.010
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr2_+_236068029
|
2.002
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr2_-_3600606
|
2.000
|
|
|
|
|
chr2_+_236067687
|
1.996
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr18_+_17575542
|
1.975
|
NM_020774
|
MIB1
|
mindbomb homolog 1 (Drosophila)
|
|
chr14_+_19458580
|
1.973
|
NM_001005483
|
OR4K5
|
olfactory receptor, family 4, subfamily K, member 5
|
|
chr2_-_200528703
|
1.973
|
NM_001039693
|
C2orf60
|
chromosome 2 open reading frame 60
|
|
chr5_+_78568094
|
1.972
|
|
|
|
|
chr11_-_9182356
|
1.957
|
|
DENND5A
|
DENN/MADD domain containing 5A
|
|
chr18_+_9904249
|
1.947
|
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr1_+_166172420
|
1.942
|
NM_001017977
NM_001198956
NM_001198957
NM_018442
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr20_-_45418818
|
1.933
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr11_+_110978309
|
1.921
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr2_-_241903787
|
1.921
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr6_+_36670117
|
1.920
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr20_+_36988368
|
1.904
|
NM_030919
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr6_-_130224108
|
1.902
|
NM_001010876
|
C6orf191
|
chromosome 6 open reading frame 191
|
|
chr1_+_159986164
|
1.902
|
NM_007240
|
DUSP12
|
dual specificity phosphatase 12
|
|
chr6_+_36670067
|
1.899
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr1_+_166172679
|
1.885
|
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr8_+_126079929
|
1.878
|
|
SQLE
|
squalene epoxidase
|
|
chr3_-_23933400
|
1.864
|
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1
|
|
chr6_+_27883877
|
1.857
|
NM_003509
|
HIST1H2AI
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H3f
|
|
chr5_+_139485703
|
1.855
|
NM_001007189
|
C5orf53
|
chromosome 5 open reading frame 53
|
|
chr2_+_241904013
|
1.852
|
|
SEPT2
|
septin 2
|
|
chr2_-_215956429
|
1.843
|
|
FN1
|
fibronectin 1
|
|
chr19_+_54068300
|
1.842
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr21_+_16024241
|
1.828
|
NM_013396
|
USP25
|
ubiquitin specific peptidase 25
|
|
chr1_+_159986217
|
1.823
|
|
DUSP12
|
dual specificity phosphatase 12
|
|
chr8_+_126079900
|
1.819
|
NM_003129
|
SQLE
|
squalene epoxidase
|
|
chr19_+_54068373
|
1.810
|
|
PPP1R15A
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr2_+_68238490
|
1.781
|
NM_020143
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr3_-_23933504
|
1.778
|
NM_020345
|
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1
|
|
chr13_-_97627491
|
1.776
|
NM_178861
|
RNF113B
|
ring finger protein 113B
|
|
chr14_+_49430109
|
1.770
|
|
ARF6
|
ADP-ribosylation factor 6
|
|
chr7_+_127079481
|
1.764
|
|
SND1
|
staphylococcal nuclease and tudor domain containing 1
|
|
chr8_+_98856984
|
1.752
|
NM_018407
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta
|
|
chr2_+_241904038
|
1.748
|
|
SEPT2
|
septin 2
|
|
chr16_+_83291107
|
1.748
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr1_+_93070120
|
1.745
|
NM_000969
|
RPL5
|
ribosomal protein L5
|
|
chr1_+_93070186
|
1.742
|
|
RPL5
|
ribosomal protein L5
|
|
chr1_+_166172545
|
1.738
|
|
DCAF6
|
DDB1 and CUL4 associated factor 6
|
|
chr6_+_12120709
|
1.737
|
NM_002114
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1
|
|
chr3_-_9414122
|
1.735
|
|
LOC440944
|
hypothetical LOC440944
|
|
chr6_+_108988797
|
1.733
|
|
FOXO3
|
forkhead box O3
|
|
chr2_+_135392879
|
1.726
|
|
CCNT2
|
cyclin T2
|
|
chr1_+_93070202
|
1.724
|
|
RPL5
|
ribosomal protein L5
|
|
chr6_+_64403936
|
1.722
|
|
PHF3
|
PHD finger protein 3
|
|
chr14_-_19553191
|
1.721
|
NM_001004712
|
OR4K14
|
olfactory receptor, family 4, subfamily K, member 14
|
|
chr7_+_106596641
|
1.720
|
NM_012257
|
HBP1
|
HMG-box transcription factor 1
|
|
chr17_-_39630951
|
1.719
|
NM_001098833
NM_020218
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr3_+_23933640
|
1.717
|
NM_002948
|
RPL15
|
ribosomal protein L15
|
|
chr18_+_53863865
|
1.716
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_+_241904024
|
1.715
|
|
SEPT2
|
septin 2
|
|
chr3_-_199161152
|
1.714
|
NM_001134435
|
IQCG
|
IQ motif containing G
|
|
chrX_+_70669672
|
1.713
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr16_+_83291097
|
1.713
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr16_+_83291055
|
1.711
|
NM_005153
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr2_+_62787534
|
1.709
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr3_+_23933618
|
1.706
|
|
RPL15
|
ribosomal protein L15
|
|
chr20_+_17897529
|
1.702
|
NM_052865
|
C20orf72
|
chromosome 20 open reading frame 72
|
|
chr6_+_15353681
|
1.699
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr2_-_241903657
|
1.695
|
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr3_+_199161446
|
1.692
|
NM_000996
|
RPL35A
|
ribosomal protein L35a
|
|
chr1_+_35507197
|
1.689
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr17_+_45221060
|
1.686
|
NM_007067
|
MYST2
|
MYST histone acetyltransferase 2
|
|
chr16_+_83291112
|
1.681
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr1_-_239865724
|
1.674
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chrX_+_70669636
|
1.674
|
NM_181672
NM_181673
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr10_-_32675848
|
1.667
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr2_+_241903395
|
1.660
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr9_+_6403321
|
1.657
|
|
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2
|
|
chr17_+_72065440
|
1.655
|
|
LOC100507246
|
hypothetical LOC100507246
|
|
chr1_-_152459506
|
1.650
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr14_+_19414230
|
1.641
|
NM_001005501
|
OR4K2
|
olfactory receptor, family 4, subfamily K, member 2
|
|
chr9_+_103200939
|
1.639
|
NM_003452
NM_197977
|
ZNF189
|
zinc finger protein 189
|
|
chr6_-_13723365
|
1.633
|
|
|
|
|
chr2_+_135392857
|
1.630
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr11_+_105453501
|
1.610
|
NM_015423
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase
|
|
chr11_+_5399916
|
1.609
|
NM_001004757
|
OR51Q1
|
olfactory receptor, family 51, subfamily Q, member 1
|
|
chr19_+_54160369
|
1.607
|
NM_000146
|
FTL
|
ferritin, light polypeptide
|
|
chr20_-_2031521
|
1.606
|
|
|
|
|
chr14_+_56805358
|
1.603
|
NM_018229
|
MUDENG
|
MU-2/AP1M2 domain containing, death-inducing
|
|
chr21_+_37661728
|
1.603
|
NM_101395
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr2_+_177785667
|
1.601
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr19_+_39611169
|
1.601
|
|
UBA2
|
ubiquitin-like modifier activating enzyme 2
|
|
chr16_+_55963899
|
1.597
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr20_+_2031418
|
1.594
|
|
STK35
|
serine/threonine kinase 35
|
|
chr6_-_37333872
|
1.585
|
NM_001162900
|
TMEM217
|
transmembrane protein 217
|
|
chr20_+_39091026
|
1.578
|
|
TOP1
|
topoisomerase (DNA) I
|
|
chr5_-_95794416
|
1.572
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr21_-_37366684
|
1.571
|
NM_153681
|
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P
|
|
chr9_+_6403183
|
1.563
|
|
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2
|
|
chr7_-_129038701
|
1.556
|
|
|
|
|
chr7_+_155129904
|
1.550
|
|
RBM33
|
RNA binding motif protein 33
|
|
chr12_+_119456538
|
1.544
|
|
RNF10
|
ring finger protein 10
|
|
chr1_+_96959762
|
1.543
|
NM_021190
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr1_-_152459667
|
1.539
|
|
C1orf43
|
chromosome 1 open reading frame 43
|
|
chr17_+_45221002
|
1.536
|
|
MYST2
|
MYST histone acetyltransferase 2
|