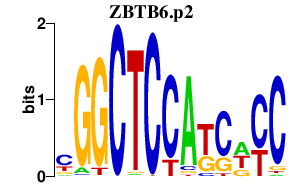
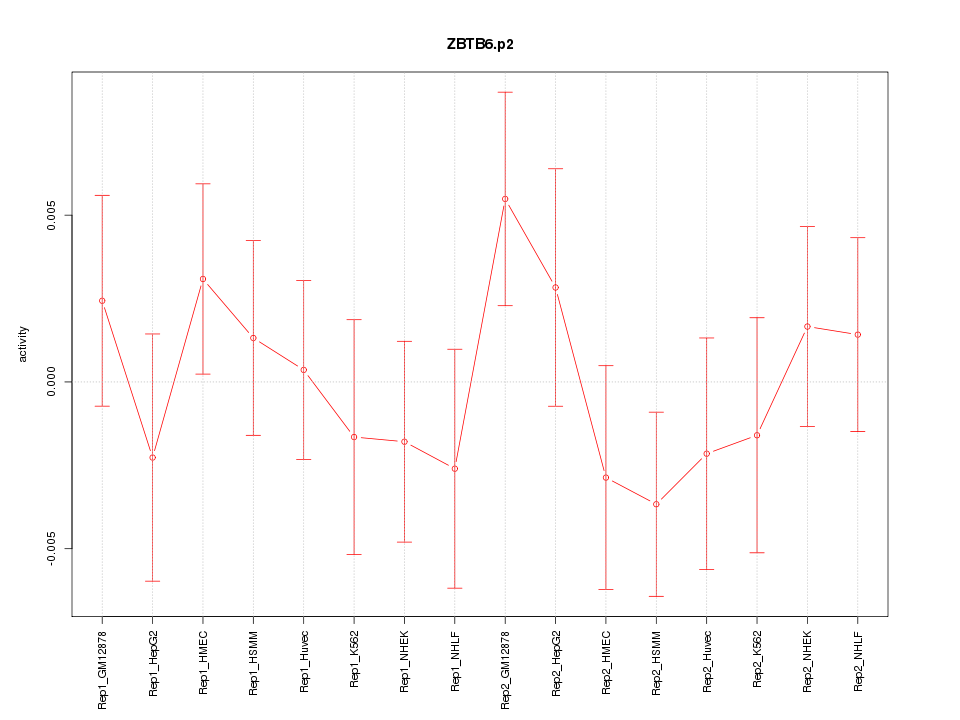
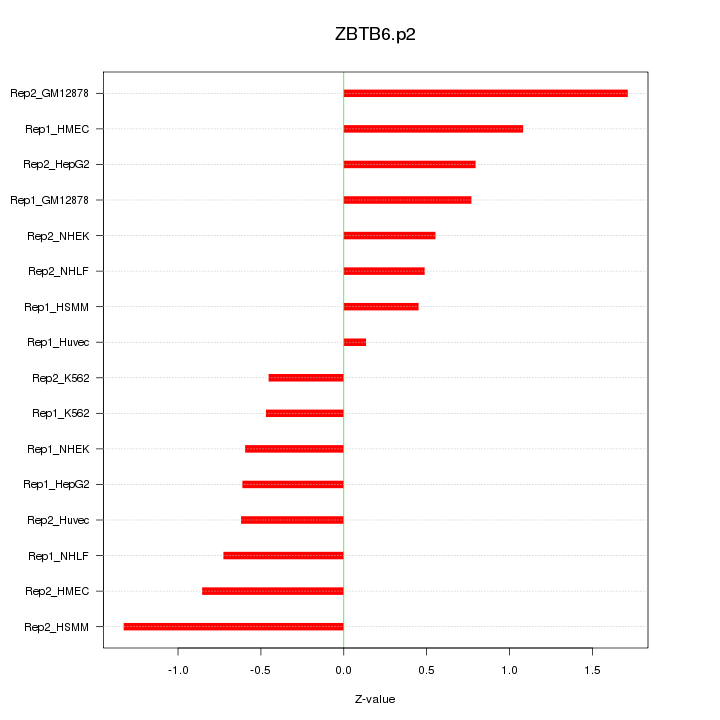
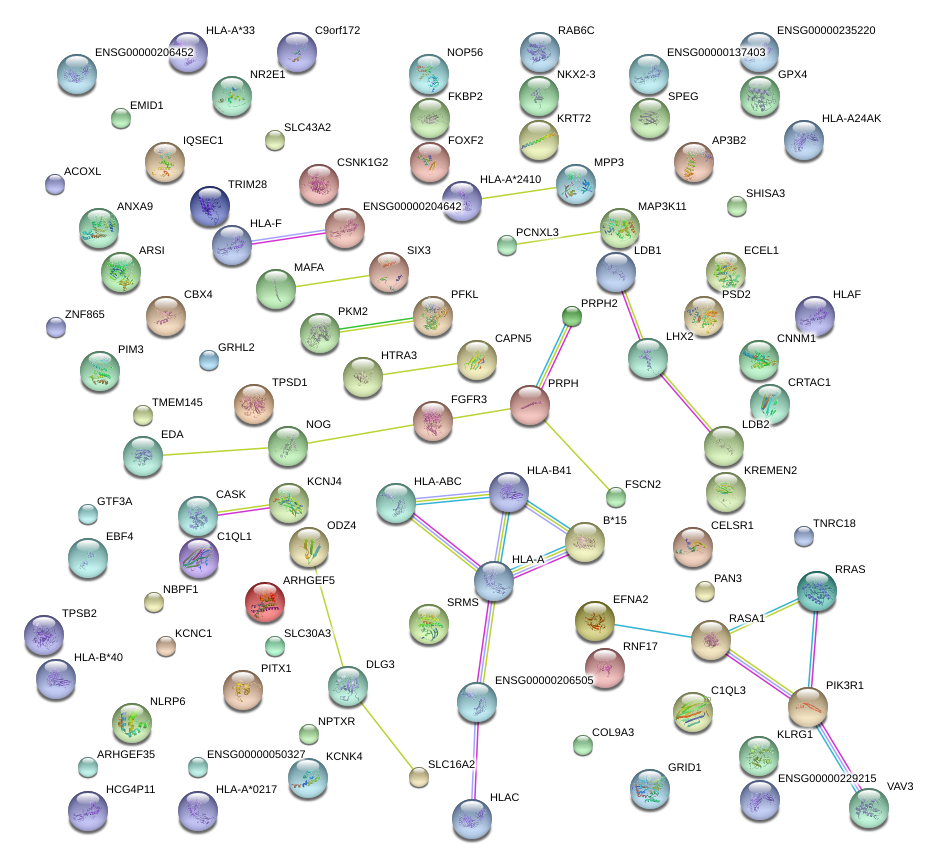
|
chr2_-_233060774
|
0.820
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr22_+_22703107
|
0.595
|
NM_001144931
|
LOC391322
|
D-dopachrome tautomerase-like
|
|
chr22_+_48740432
|
0.583
|
|
PIM3
|
pim-3 oncogene
|
|
chr9_+_125813849
|
0.547
|
|
LHX2
|
LIM homeobox 2
|
|
chr10_-_88116181
|
0.508
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr20_-_61649259
|
0.504
|
NM_080823
|
SRMS
|
src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites
|
|
chr10_+_101282679
|
0.499
|
NM_145285
|
NKX2-3
|
NK2 transcription factor related, locus 3 (Drosophila)
|
|
chr22_-_45311730
|
0.499
|
NM_014246
|
CELSR1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr2_+_220017617
|
0.487
|
|
SPEG
|
SPEG complex locus
|
|
chr13_+_27610379
|
0.480
|
NM_175854
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr7_-_5429702
|
0.461
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr2_+_111206620
|
0.450
|
NM_001142807
|
ACOXL
|
acyl-CoA oxidase-like
|
|
chr7_+_143683365
|
0.449
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr22_-_37569962
|
0.449
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr17_-_40401138
|
0.447
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr4_+_1764802
|
0.446
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr6_+_29799182
|
0.439
|
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr20_+_2581177
|
0.439
|
NM_006392
|
NOP56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr17_-_75427644
|
0.432
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr6_+_29799095
|
0.419
|
NM_001098478
NM_001098479
NM_018950
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr20_+_60918836
|
0.381
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr17_+_77110011
|
0.381
|
NM_001077182
NM_012418
|
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus)
|
|
chr5_-_141238011
|
0.380
|
|
|
|
|
chr11_-_65139277
|
0.378
|
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr11_-_78829342
|
0.376
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr5_+_139155569
|
0.375
|
NM_032289
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr1_+_149221122
|
0.366
|
NM_003568
|
ANXA9
|
annexin A9
|
|
chr19_+_60816770
|
0.361
|
NM_001195605
|
ZNF865
|
zinc finger protein 865
|
|
chr9_+_138858687
|
0.338
|
NM_001080482
|
C9orf172
|
chromosome 9 open reading frame 172
|
|
chrX_-_41667174
|
0.335
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr16_+_2954217
|
0.334
|
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr5_+_67547357
|
0.331
|
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chrX_+_68752635
|
0.327
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr10_-_103869923
|
0.323
|
NM_001113407
|
LDB1
|
LIM domain binding 1
|
|
chr3_-_12983959
|
0.319
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr6_+_108593907
|
0.316
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr2_-_131838200
|
0.316
|
NM_001077637
|
LOC150786
|
RAB6C-like
|
|
chrX_+_49531209
|
0.313
|
NM_001145073
|
USP27X
|
ubiquitin specific peptidase 27, X-linked
|
|
chr15_-_81175542
|
0.301
|
NM_004644
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr19_+_1055614
|
0.298
|
NM_001039848
|
GPX4
|
glutathione peroxidase 4 (phospholipid hydroperoxidase)
|
|
chrX_+_73557809
|
0.292
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr6_+_1335067
|
0.292
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr11_+_17714070
|
0.288
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr22_-_37180958
|
0.278
|
NM_152868
|
KCNJ4
|
potassium inwardly-rectifying channel, subfamily J, member 4
|
|
chr5_-_134397740
|
0.278
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr16_+_65018316
|
0.273
|
NM_001136106
NM_001178020
NM_001197224
NM_001197225
|
BEAN1
|
brain expressed, associated with NEDD4, 1
|
|
chr19_+_47509274
|
0.271
|
NM_173633
|
TMEM145
|
transmembrane protein 145
|
|
chr13_+_26896614
|
0.270
|
NM_002097
|
GTF3A
|
general transcription factor IIIA
|
|
chr6_+_30018287
|
0.268
|
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr2_+_45022540
|
0.267
|
NM_005413
|
SIX3
|
SIX homeobox 3
|
|
chr21_+_44549486
|
0.267
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr11_+_75056343
|
0.267
|
|
|
|
|
chr20_+_2621193
|
0.265
|
NM_001110514
|
EBF4
|
early B-cell factor 4
|
|
chr5_-_149662640
|
0.265
|
NM_001012301
|
ARSI
|
arylsulfatase family, member I
|
|
chr10_-_99780574
|
0.264
|
NM_018058
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr11_+_65140207
|
0.263
|
NM_032223
|
PCNXL3
|
pecanex-like 3 (Drosophila)
|
|
chr22_+_27931685
|
0.261
|
NM_133455
|
EMID1
|
EMI domain containing 1
|
|
chr11_+_63815348
|
0.260
|
NM_033310
|
KCNK4
|
potassium channel, subfamily K, member 4
|
|
chr19_+_63747829
|
0.259
|
|
TRIM28
|
tripartite motif containing 28
|
|
chr4_+_42094612
|
0.258
|
NM_001080505
|
SHISA3
|
shisa homolog 3 (Xenopus laevis)
|
|
chr12_-_51281482
|
0.258
|
NM_001146225
NM_001146226
NM_080747
|
KRT72
|
keratin 72
|
|
chr8_-_144583718
|
0.258
|
NM_201589
|
MAFA
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian)
|
|
chr9_+_125813672
|
0.256
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chr15_-_70288071
|
0.254
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chrX_+_69581429
|
0.253
|
NM_021120
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr2_-_27339278
|
0.253
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr6_+_30018265
|
0.252
|
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr17_-_1478293
|
0.251
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr12_+_47975168
|
0.250
|
NM_006262
|
PRPH
|
peripherin
|
|
chr16_+_1246273
|
0.250
|
NM_012217
|
TPSD1
|
tryptase delta 1
|
|
chr19_+_1892157
|
0.247
|
NM_001319
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr17_+_52026058
|
0.244
|
NM_005450
|
NOG
|
noggin
|
|
chr19_+_1236766
|
0.243
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr11_+_63765106
|
0.242
|
NM_001135208
|
FKBP2
|
FK506 binding protein 2, 13kDa
|
|
chr10_+_101078784
|
0.242
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chr1_-_108308997
|
0.242
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr4_+_8322358
|
0.241
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr19_+_63747638
|
0.240
|
NM_005762
|
TRIM28
|
tripartite motif containing 28
|
|
chr17_+_4434582
|
0.239
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr1_+_12108542
|
0.238
|
NM_152942
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr1_-_2451543
|
0.238
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr17_+_7549138
|
0.238
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr16_+_54782802
|
0.238
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr17_+_27617307
|
0.237
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr15_+_39573363
|
0.236
|
NM_002220
|
ITPKA
|
inositol 1,4,5-trisphosphate 3-kinase A
|
|
chr12_+_52634950
|
0.234
|
NM_173860
|
HOXC12
|
homeobox C12
|
|
chr1_-_226530350
|
0.234
|
|
|
|
|
chr16_-_85099941
|
0.233
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr7_-_4965270
|
0.233
|
NM_001100600
NM_198403
|
MMD2
|
monocyte to macrophage differentiation-associated 2
|
|
chrX_+_49573952
|
0.233
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr17_+_73638430
|
0.233
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr19_+_3149813
|
0.231
|
|
NCLN
|
nicalin
|
|
chr22_-_35829402
|
0.230
|
NM_153609
|
TMPRSS6
|
transmembrane protease, serine 6
|
|
chr22_-_37569679
|
0.230
|
|
|
|
|
chr1_+_2149993
|
0.230
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr5_-_141238127
|
0.229
|
NM_002587
NM_032420
|
PCDH1
|
protocadherin 1
|
|
chr1_-_8406288
|
0.228
|
NM_001042682
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats
|
|
chr16_+_28742066
|
0.228
|
|
ATXN2L
|
ataxin 2-like
|
|
chr10_+_103815114
|
0.227
|
NM_024747
|
HPS6
|
Hermansky-Pudlak syndrome 6
|
|
chr8_+_126511739
|
0.225
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr19_+_10843369
|
0.225
|
|
CARM1
|
coactivator-associated arginine methyltransferase 1
|
|
chr11_+_268569
|
0.223
|
NM_138329
|
NLRP6
|
NLR family, pyrin domain containing 6
|
|
chr3_+_130203161
|
0.223
|
NM_024768
|
CCDC48
|
coiled-coil domain containing 48
|
|
chr2_-_232354067
|
0.222
|
NM_002601
|
PDE6D
|
phosphodiesterase 6D, cGMP-specific, rod, delta
|
|
chr22_+_38246509
|
0.221
|
NM_001675
NM_182810
|
ATF4
|
activating transcription factor 4 (tax-responsive enhancer element B67)
|
|
chr19_+_44697592
|
0.221
|
NM_182704
|
SELV
|
selenoprotein V
|
|
chr16_+_54782751
|
0.221
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr19_+_59333287
|
0.221
|
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr19_-_40693176
|
0.220
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr20_+_56861430
|
0.220
|
NM_001077490
NM_080425
|
GNAS
|
GNAS complex locus
|
|
chr8_-_81104938
|
0.219
|
NM_014018
|
MRPS28
|
mitochondrial ribosomal protein S28
|
|
chr6_+_33352886
|
0.218
|
NM_003782
|
B3GALT4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4
|
|
chr2_+_233448739
|
0.218
|
|
C2orf82
|
chromosome 2 open reading frame 82
|
|
chr11_+_67567022
|
0.217
|
NM_006053
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr20_-_4177519
|
0.215
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr16_+_65754788
|
0.215
|
NM_001040667
NM_001538
|
HSF4
|
heat shock transcription factor 4
|
|
chr22_-_49240594
|
0.215
|
|
SBF1
|
SET binding factor 1
|
|
chr2_+_26840626
|
0.214
|
NM_017877
|
CENPA
C2orf18
|
centromere protein A
chromosome 2 open reading frame 18
|
|
chr17_-_77474597
|
0.214
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr12_-_6680053
|
0.212
|
NM_153685
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
chrX_-_54226364
|
0.211
|
NM_017848
NM_198456
|
FAM120C
|
family with sequence similarity 120C
|
|
chr14_-_22947320
|
0.211
|
NM_002471
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha
|
|
chrX_-_50230329
|
0.211
|
NM_001013742
|
DGKK
|
diacylglycerol kinase, kappa
|
|
chr5_+_10617431
|
0.210
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr21_+_44169706
|
0.209
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr22_-_22252376
|
0.209
|
NM_020070
NM_152855
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1
|
|
chr22_+_29819343
|
0.208
|
|
SMTN
|
smoothelin
|
|
chr1_+_149437602
|
0.208
|
NM_001135636
NM_001135637
NM_001135638
NM_003557
|
PIP5K1A
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha
|
|
chr16_+_11669736
|
0.206
|
NM_003498
|
SNN
|
stannin
|
|
chr7_+_7974941
|
0.205
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr2_+_231968578
|
0.205
|
NM_145236
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7
|
|
chr19_+_62703113
|
0.204
|
NM_198542
|
ZNF773
|
zinc finger protein 773
|
|
chr16_-_31007630
|
0.204
|
NM_001039503
|
PRSS53
|
protease, serine, 53
|
|
chr7_+_154943466
|
0.203
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr20_-_60228665
|
0.203
|
NM_007232
|
HRH3
|
histamine receptor H3
|
|
chr10_+_134060681
|
0.203
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr12_+_52680143
|
0.202
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr8_-_145187553
|
0.201
|
NM_017570
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing)
|
|
chr10_+_99069007
|
0.201
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr16_-_65984836
|
0.199
|
NM_015964
NM_016140
|
TPPP3
|
tubulin polymerization-promoting protein family member 3
|
|
chr11_+_63764988
|
0.199
|
NM_004470
|
FKBP2
|
FK506 binding protein 2, 13kDa
|
|
chr17_-_76985538
|
0.199
|
|
|
|
|
chrX_-_39841599
|
0.198
|
NM_001123385
NM_017745
|
BCOR
|
BCL6 corepressor
|
|
chr14_-_87859284
|
0.198
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr19_+_63747678
|
0.197
|
|
TRIM28
|
tripartite motif containing 28
|
|
chr2_+_101235695
|
0.195
|
NM_017546
|
C2orf29
|
chromosome 2 open reading frame 29
|
|
chr21_+_44569537
|
0.194
|
|
PFKL
|
phosphofructokinase, liver
|
|
chr17_-_44663107
|
0.194
|
NM_001143804
NM_178500
|
PHOSPHO1
|
phosphatase, orphan 1
|
|
chr14_+_103675194
|
0.193
|
|
KIF26A
|
kinesin family member 26A
|
|
chr2_+_220200530
|
0.193
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chr1_+_109594163
|
0.193
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr19_+_59333232
|
0.192
|
NM_014516
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr6_+_1555679
|
0.192
|
NM_001453
|
FOXC1
|
forkhead box C1
|
|
chrX_+_12066505
|
0.192
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr17_+_4348877
|
0.191
|
NM_001124758
|
SPNS2
|
spinster homolog 2 (Drosophila)
|
|
chr17_+_46067114
|
0.191
|
NM_001144070
NM_003786
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3
|
|
chr17_+_32032113
|
0.190
|
NM_024864
|
MRM1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr15_-_38420414
|
0.190
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr20_-_61574259
|
0.190
|
NM_004518
NM_172106
NM_172107
NM_172108
NM_172109
|
KCNQ2
|
potassium voltage-gated channel, KQT-like subfamily, member 2
|
|
chr14_+_102662416
|
0.190
|
NM_006291
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr22_-_45512708
|
0.190
|
|
CERK
|
ceramide kinase
|
|
chr22_+_36384647
|
0.189
|
NM_020315
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr22_+_38720881
|
0.187
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr3_+_48482625
|
0.186
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr12_+_131797455
|
0.186
|
NM_001170543
NM_001170544
NM_138575
|
PGAM5
|
phosphoglycerate mutase family member 5
|
|
chr11_-_75056761
|
0.186
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr1_+_41022066
|
0.185
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr22_-_41419800
|
0.183
|
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr12_+_439803
|
0.183
|
NM_173593
|
B4GALNT3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3
|
|
chr20_+_57309042
|
0.182
|
|
EDN3
|
endothelin 3
|
|
chr19_-_56612653
|
0.182
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr22_+_37431900
|
0.182
|
|
GTPBP1
|
GTP binding protein 1
|
|
chr5_+_175231144
|
0.180
|
|
CPLX2
|
complexin 2
|
|
chr12_+_50686990
|
0.179
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr8_+_145461356
|
0.179
|
NM_001008271
NM_001080514
|
SCXA
SCXB
|
scleraxis homolog A (mouse)
scleraxis homolog B (mouse)
|
|
chr5_+_168996852
|
0.179
|
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr10_-_121286034
|
0.178
|
NM_002925
|
RGS10
|
regulator of G-protein signaling 10
|
|
chr16_-_21439195
|
0.178
|
|
SLC7A5P2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 pseudogene 2
|
|
chr19_+_2115119
|
0.177
|
NM_032482
|
DOT1L
|
DOT1-like, histone H3 methyltransferase (S. cerevisiae)
|
|
chr7_-_1465487
|
0.177
|
|
MICALL2
|
MICAL-like 2
|
|
chr3_+_10832867
|
0.177
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr5_+_168996816
|
0.177
|
NM_004946
|
DOCK2
|
dedicator of cytokinesis 2
|
|
chr6_+_1556543
|
0.177
|
|
FOXC1
|
forkhead box C1
|
|
chr9_+_133338775
|
0.177
|
|
PRRC2B
|
proline-rich coiled-coil 2B
|
|
chr16_-_2099854
|
0.176
|
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr19_+_3136965
|
0.176
|
|
NCLN
|
nicalin
|
|
chr8_-_21702273
|
0.176
|
NM_001165038
NM_001165039
NM_001495
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr17_+_8865547
|
0.174
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr19_+_1892184
|
0.174
|
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr2_+_219432789
|
0.174
|
NM_006522
|
WNT6
|
wingless-type MMTV integration site family, member 6
|
|
chr13_+_112681626
|
0.174
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr22_-_48835181
|
0.173
|
NM_001080447
|
TTLL8
|
tubulin tyrosine ligase-like family, member 8
|
|
chr16_-_28411123
|
0.173
|
NM_001042432
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr3_+_182912405
|
0.173
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr1_-_13712744
|
0.173
|
NM_001010847
|
LRRC38
|
leucine rich repeat containing 38
|
|
chr12_+_48737753
|
0.172
|
NM_001095
NM_020039
|
ACCN2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr8_+_27404508
|
0.172
|
NM_001979
|
EPHX2
|
epoxide hydrolase 2, cytoplasmic
|
|
chr12_+_109956210
|
0.172
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr19_+_46390980
|
0.172
|
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr19_+_55844446
|
0.172
|
NM_001195076
|
LOC342918
|
hypothetical protein LOC342918
|