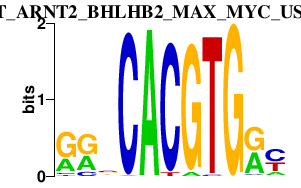
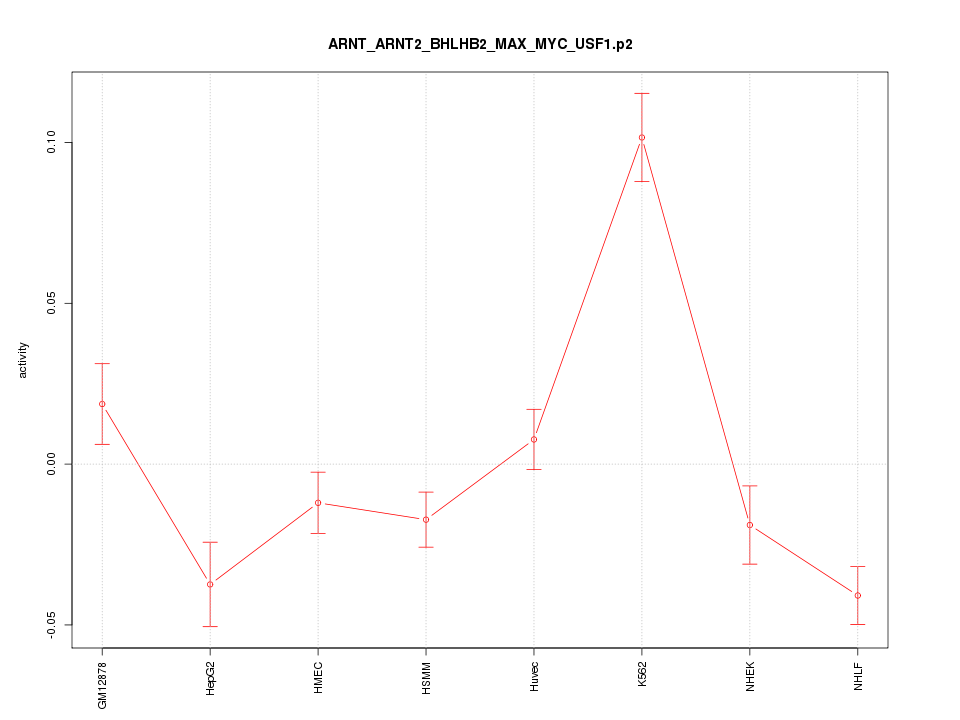
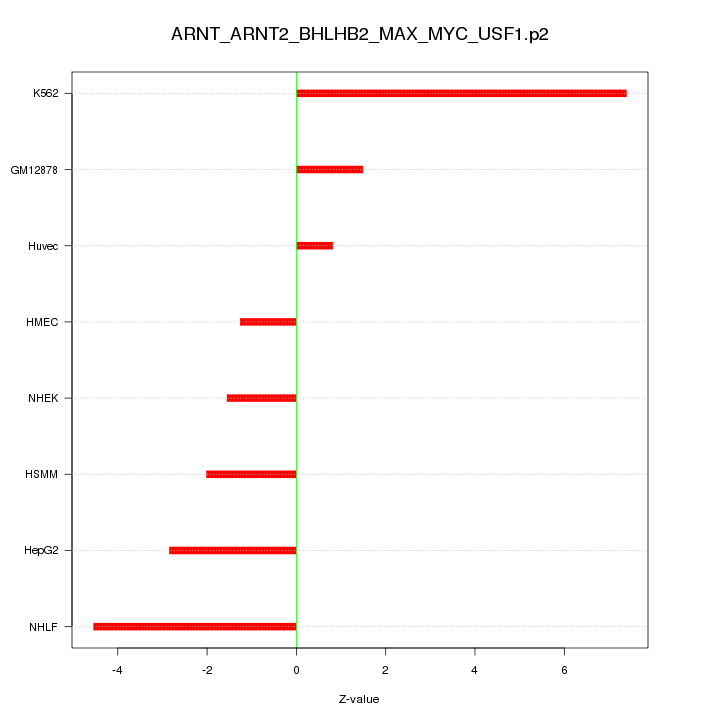
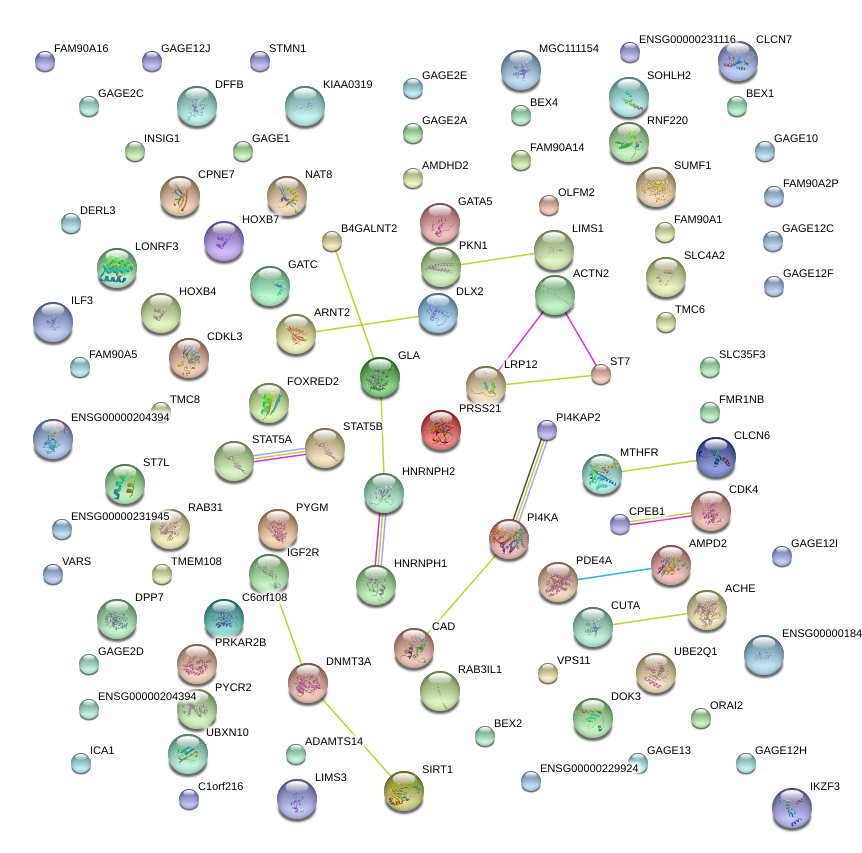
|
chr17_-_53764867
|
8.846
|
|
|
|
|
chr16_+_2510323
|
7.071
|
NM_001145815
NM_015944
|
AMDHD2
|
amidohydrolase domain containing 2
|
|
chr1_+_20385164
|
7.059
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr7_+_150390566
|
6.907
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr16_-_1465021
|
5.917
|
|
CLCN7
|
chloride channel 7
|
|
chr16_-_1465007
|
4.948
|
|
CLCN7
|
chloride channel 7
|
|
chr12_-_56432292
|
4.898
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_-_152797820
|
4.750
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr11_-_61441536
|
4.689
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr12_-_56432377
|
4.557
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr12_-_56432375
|
4.486
|
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr1_-_152797760
|
4.476
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr1_-_152797707
|
4.348
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr9_-_139128777
|
4.318
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr1_-_152797767
|
4.276
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr17_-_44010543
|
4.252
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr16_-_1465041
|
4.222
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr12_-_56432385
|
4.156
|
NM_000075
|
CDK4
|
cyclin-dependent kinase 4
|
|
chr16_-_1464927
|
3.895
|
|
CLCN7
|
chloride channel 7
|
|
chr1_-_11788666
|
3.725
|
NM_005957
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr9_-_139128983
|
3.672
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr8_+_142471290
|
3.670
|
|
|
|
|
chr17_-_44043361
|
3.652
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr17_-_73636362
|
3.626
|
|
TMC6
|
transmembrane channel-like 6
|
|
chrX_-_100549444
|
3.558
|
|
GLA
|
galactosidase, alpha
|
|
chr19_+_10626106
|
3.556
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr17_-_44043278
|
3.532
|
|
HOXB7
|
homeobox B7
|
|
chrX_+_100549885
|
3.507
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chrX_+_100549869
|
3.500
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr1_-_11788550
|
3.369
|
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr1_+_234916376
|
3.269
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr20_-_60484420
|
3.192
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chr11_-_71491942
|
3.141
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr10_+_72102563
|
3.131
|
NM_080722
NM_139155
|
ADAMTS14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14
|
|
chr1_-_35957030
|
3.120
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chrX_+_100549861
|
3.076
|
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr22_-_35233032
|
3.062
|
NM_024955
NM_001102371
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr2_+_108571177
|
2.987
|
NM_004987
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr11_-_71491983
|
2.938
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chrX_+_100549839
|
2.900
|
NM_001032393
NM_019597
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H')
|
|
chr19_+_10392128
|
2.878
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr3_-_4483952
|
2.869
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr17_-_44042924
|
2.847
|
|
HOXB7
|
homeobox B7
|
|
chr22_-_35232979
|
2.842
|
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr16_+_88169612
|
2.757
|
NM_014427
NM_153636
|
CPNE7
|
copine VII
|
|
chr6_-_43305156
|
2.754
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chrX_-_100549561
|
2.721
|
NM_000169
|
GLA
|
galactosidase, alpha
|
|
chr1_-_35956979
|
2.718
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr11_-_71492019
|
2.685
|
NM_017907
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr2_+_27293846
|
2.675
|
|
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
|
|
chr11_-_71491916
|
2.664
|
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1
|
|
chr6_+_160310118
|
2.652
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr1_+_11788889
|
2.627
|
|
CLCN6
|
chloride channel 6
|
|
chr3_-_4483928
|
2.579
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr17_-_73636386
|
2.566
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chrX_+_117992603
|
2.555
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr22_-_22511173
|
2.551
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chr1_-_224178600
|
2.522
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr6_-_33493792
|
2.511
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr3_-_4483955
|
2.497
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr7_+_116380590
|
2.490
|
NM_018412
NM_021908
|
ST7
|
suppression of tumorigenicity 7
|
|
chrX_+_102356659
|
2.451
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chr2_+_108571209
|
2.450
|
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr5_-_133734616
|
2.437
|
|
CDKL3
|
cyclin-dependent kinase-like 3
|
|
chr17_-_35273947
|
2.428
|
NM_012481
NM_183228
NM_183229
NM_183230
NM_183231
NM_183232
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos)
|
|
chr18_+_9698299
|
2.372
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr7_+_154720416
|
2.367
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chrX_+_117992623
|
2.348
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr12_-_8271460
|
2.346
|
NM_018088
|
FAM90A1
|
family with sequence similarity 90, member A1
|
|
chr19_-_9908014
|
2.335
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr1_-_26105954
|
2.333
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr16_+_2807164
|
2.328
|
NM_006799
NM_144956
NM_144957
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr1_+_44870256
|
2.320
|
|
RNF220
|
ring finger protein 220
|
|
chr3_+_134239860
|
2.304
|
NM_001136469
NM_023943
|
TMEM108
|
transmembrane protein 108
|
|
chr22_-_19543099
|
2.304
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr2_-_25328452
|
2.289
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr1_+_44870223
|
2.272
|
|
RNF220
|
ring finger protein 220
|
|
chr5_-_176869943
|
2.249
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr6_-_31871394
|
2.240
|
|
VARS
|
valyl-tRNA synthetase
|
|
chr11_+_118443683
|
2.229
|
NM_021729
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chrX_-_102452461
|
2.228
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr16_+_2807228
|
2.221
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chrX_+_49241061
|
2.218
|
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 2A
|
|
chr15_-_81113676
|
2.214
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chrX_+_146870540
|
2.202
|
NM_152578
|
FMR1NB
|
fragile X mental retardation 1 neighbor
|
|
chr7_-_100331417
|
2.198
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr1_-_224178606
|
2.198
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr9_-_139128979
|
2.194
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr2_-_172675873
|
2.193
|
|
DLX2
|
distal-less homeobox 2
|
|
chr17_+_37694007
|
2.178
|
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr17_+_73638430
|
2.178
|
NM_152468
|
TMC8
|
transmembrane channel-like 8
|
|
chr19_+_14405100
|
2.169
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr1_+_109963950
|
2.147
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr9_-_139129015
|
2.141
|
NM_013379
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr10_+_69314714
|
2.130
|
|
SIRT1
|
sirtuin 1
|
|
chr3_-_4483939
|
2.122
|
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr7_-_8268206
|
2.120
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr18_+_9698269
|
2.113
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr7_+_101860958
|
2.099
|
NM_001126340
NM_032831
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr7_-_8268292
|
2.092
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr11_+_118443740
|
2.092
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr7_+_106472243
|
2.088
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr22_-_19543012
|
2.078
|
|
PI4KA
|
phosphatidylinositol 4-kinase, catalytic, alpha
|
|
chr9_-_139128992
|
2.070
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr13_-_35686665
|
2.053
|
NM_017826
|
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2
|
|
chr1_-_40335451
|
2.043
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr11_+_118443738
|
2.031
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr19_+_14405216
|
2.014
|
|
PKN1
|
protein kinase N1
|
|
chr7_+_116380790
|
2.013
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr17_-_37586716
|
2.013
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr7_+_50314801
|
2.011
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr1_-_224178553
|
2.008
|
NM_013328
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr1_-_40335496
|
2.003
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr12_-_107551783
|
1.992
|
NM_003006
|
SELPLG
|
selectin P ligand
|
|
chr12_+_54396118
|
1.983
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr19_-_6375804
|
1.979
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr1_-_52936573
|
1.975
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr19_+_10625987
|
1.972
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr11_+_118443725
|
1.970
|
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae)
|
|
chr9_-_139128990
|
1.967
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr1_-_26105477
|
1.964
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr19_-_53516962
|
1.959
|
|
CCDC114
|
coiled-coil domain containing 114
|
|
chr11_+_35641234
|
1.955
|
|
TRIM44
|
tripartite motif containing 44
|
|
chrX_+_49075036
|
1.952
|
NM_012196
NM_001127200
NM_001098412
|
GAGE8
GAGE2C
GAGE2E
GAGE13
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 13
G antigen 2A
|
|
chr12_-_107551694
|
1.947
|
|
SELPLG
|
selectin P ligand
|
|
chr12_+_54396099
|
1.942
|
|
BLOC1S1-RDH5
BLOC1S1
|
BLOC1S1-RDH5 read-through transcript
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr7_-_8268666
|
1.934
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr1_-_224178486
|
1.933
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr7_+_101861007
|
1.929
|
|
ORAI2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr3_+_134239906
|
1.929
|
|
TMEM108
|
transmembrane protein 108
|
|
chr22_+_18485145
|
1.925
|
|
RANBP1
|
RAN binding protein 1
|
|
chr12_+_54396086
|
1.916
|
NM_001487
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr9_-_139128988
|
1.913
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr12_-_107551744
|
1.910
|
|
SELPLG
|
selectin P ligand
|
|
chr6_-_33493898
|
1.907
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr7_+_100302885
|
1.902
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr10_-_45410197
|
1.895
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr7_+_154720547
|
1.888
|
|
INSIG1
|
insulin induced gene 1
|
|
chr22_+_18485057
|
1.879
|
|
RANBP1
|
RAN binding protein 1
|
|
chr1_+_11788827
|
1.872
|
|
CLCN6
|
chloride channel 6
|
|
chr1_+_109963982
|
1.868
|
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr1_+_32992131
|
1.858
|
|
|
|
|
chr22_+_18485076
|
1.856
|
|
RANBP1
|
RAN binding protein 1
|
|
chr1_-_40335510
|
1.854
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr2_-_172572993
|
1.847
|
|
|
|
|
chr1_-_40335504
|
1.847
|
|
PPT1
|
palmitoyl-protein thioesterase 1
|
|
chr22_-_19180075
|
1.840
|
NM_032775
|
KLHL22
|
kelch-like 22 (Drosophila)
|
|
chr17_+_37941476
|
1.833
|
NM_000263
|
NAGLU
|
N-acetylglucosaminidase, alpha
|
|
chr22_-_17799177
|
1.833
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr1_-_27099515
|
1.833
|
|
GPATCH3
|
G patch domain containing 3
|
|
chr1_-_21868380
|
1.828
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr3_+_160002582
|
1.803
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr1_-_11042668
|
1.790
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr1_+_11788793
|
1.784
|
NM_001286
NM_021735
NM_021736
NM_021737
|
CLCN6
|
chloride channel 6
|
|
chr1_+_26609819
|
1.772
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr22_+_18485068
|
1.765
|
|
RANBP1
|
RAN binding protein 1
|
|
chr3_+_160002605
|
1.762
|
|
MFSD1
|
major facilitator superfamily domain containing 1
|
|
chr1_+_1157485
|
1.756
|
NM_080605
|
B3GALT6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6
|
|
chr16_+_80370426
|
1.739
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr7_-_150305933
|
1.731
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr19_-_6375782
|
1.718
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr17_+_63252087
|
1.713
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr10_-_50066386
|
1.691
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr17_-_44037332
|
1.684
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr4_-_90197140
|
1.663
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr2_-_120697148
|
1.662
|
|
TMEM185B
|
transmembrane protein 185B (pseudogene)
|
|
chr19_-_6375820
|
1.660
|
NM_003685
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr22_-_17798998
|
1.657
|
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chr7_+_149696792
|
1.655
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr8_-_66863841
|
1.651
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr22_+_18484810
|
1.650
|
NM_002882
|
RANBP1
|
RAN binding protein 1
|
|
chr1_-_224178475
|
1.650
|
|
PYCR2
|
pyrroline-5-carboxylate reductase family, member 2
|
|
chr18_+_9698187
|
1.649
|
NM_006868
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr17_-_43977273
|
1.648
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr10_+_69314347
|
1.645
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr19_+_1018536
|
1.644
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr16_-_87570716
|
1.632
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr9_+_130124566
|
1.625
|
NM_016035
|
COQ4
|
coenzyme Q4 homolog (S. cerevisiae)
|
|
chr3_+_185515108
|
1.618
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr1_+_42920946
|
1.616
|
|
YBX1
|
Y box binding protein 1
|
|
chr12_-_46439127
|
1.604
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr1_-_201162938
|
1.604
|
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chrX_+_55263515
|
1.603
|
NM_130467
NM_001013435
|
PAGE5
|
P antigen family, member 5 (prostate associated)
|
|
chr22_-_17799215
|
1.593
|
NM_003325
|
HIRA
|
HIR histone cell cycle regulation defective homolog A (S. cerevisiae)
|
|
chrX_+_134482387
|
1.592
|
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr1_-_52936602
|
1.582
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr1_-_1700088
|
1.566
|
NM_001198993
|
NADK
|
NAD kinase
|
|
chr2_+_26249463
|
1.553
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr19_+_748390
|
1.552
|
NM_002819
NM_031990
NM_031991
NM_175847
|
PTBP1
|
polypyrimidine tract binding protein 1
|
|
chrX_+_151991474
|
1.552
|
NM_001170944
NM_032882
|
LOC100287428
PNMA6A
|
paraneoplastic antigen-like protein 6A related gene
paraneoplastic antigen like 6A
|
|
chr14_-_64416307
|
1.549
|
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr15_-_48766252
|
1.548
|
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr2_-_80384454
|
1.548
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr18_+_9698322
|
1.546
|
|
RAB31
|
RAB31, member RAS oncogene family
|
|
chr20_+_60744231
|
1.543
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chrX_+_49094059
|
1.543
|
NM_012196
NM_001472
NM_001127200
NM_001098407
NM_001127212
|
GAGE8
GAGE2C
GAGE2E
GAGE2D
GAGE2A
|
G antigen 8
G antigen 2C
G antigen 2E
G antigen 2D
G antigen 2A
|
|
chr19_-_4509471
|
1.540
|
NM_032108
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chrX_+_134482212
|
1.539
|
NM_182540
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B
|
|
chr9_-_139316523
|
1.531
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr5_+_133734768
|
1.528
|
NM_003337
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|