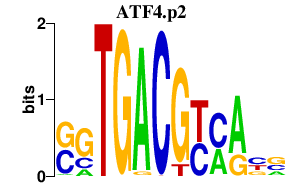
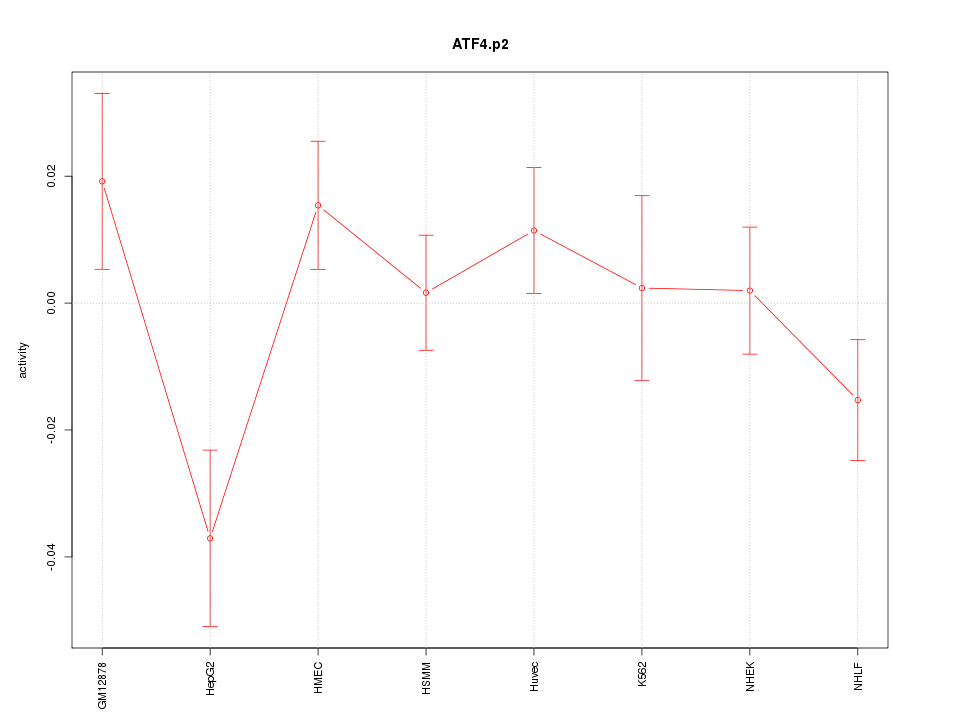
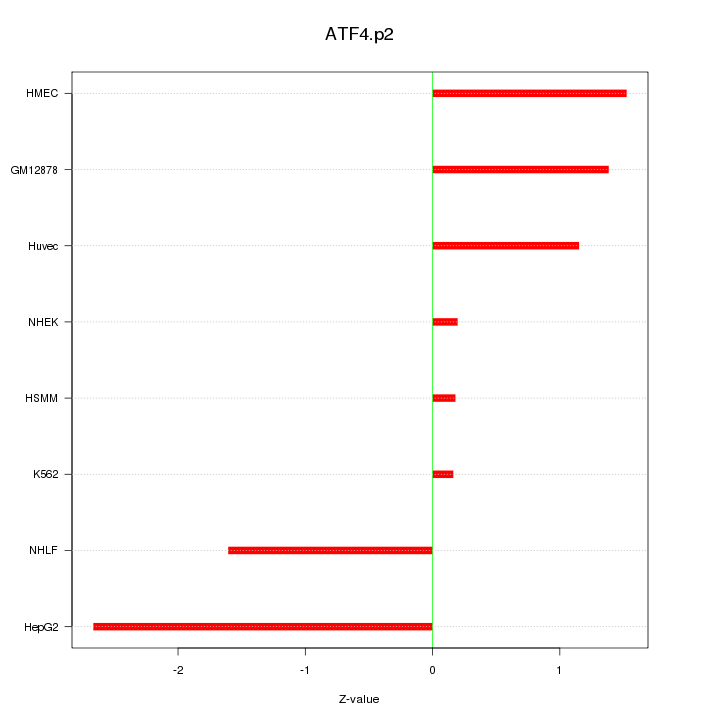
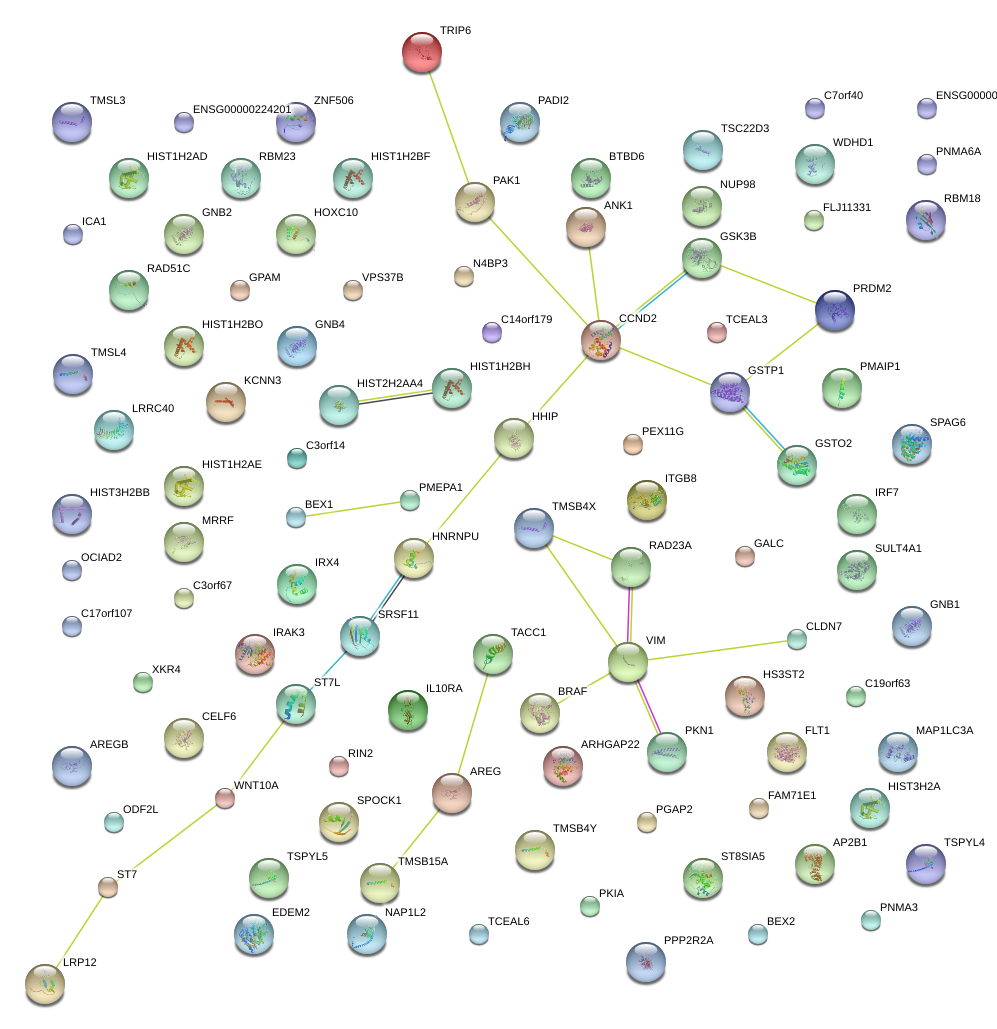
|
chr20_+_32610161
|
5.191
|
NM_032514
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha
|
|
chr7_-_44992729
|
3.365
|
|
C7orf40
|
chromosome 7 open reading frame 40
|
|
chr19_+_55671600
|
1.599
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr19_+_55671594
|
1.533
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr19_+_55671468
|
1.523
|
NM_175063
NM_206538
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr19_+_55671584
|
1.523
|
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr19_-_55671273
|
1.329
|
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr14_+_75521806
|
1.301
|
NM_001102564
NM_052873
|
C14orf179
|
chromosome 14 open reading frame 179
|
|
chr1_+_13899272
|
1.236
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr10_-_113933457
|
1.232
|
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr4_-_48603501
|
1.190
|
NM_001014446
NM_152398
|
OCIAD2
|
OCIA domain containing 2
|
|
chr19_-_7459867
|
1.188
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr10_+_22674379
|
1.164
|
NM_012443
NM_172242
|
SPAG6
|
sperm associated antigen 6
|
|
chr20_+_19863716
|
1.156
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr4_+_75529716
|
1.142
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr1_-_243092581
|
1.133
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr12_+_52665196
|
1.109
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr11_+_67107852
|
1.083
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr2_+_219453487
|
1.066
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr16_+_22732982
|
1.048
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr10_+_106018620
|
1.046
|
NM_001191013
NM_183239
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chr8_+_38763878
|
1.041
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr20_-_33329319
|
1.026
|
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr20_-_55718351
|
1.022
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr7_+_100302885
|
0.997
|
NM_003302
|
TRIP6
|
thyroid hormone receptor interactor 6
|
|
chr1_-_86634511
|
0.990
|
NM_001007022
NM_001184765
NM_001184766
NM_020729
|
ODF2L
|
outer dense fiber of sperm tails 2-like
|
|
chrX_+_12903145
|
0.973
|
NM_021109
NM_183049
|
TMSB4X
TMSL3
|
thymosin beta 4, X-linked
thymosin-like 3
|
|
chr18_-_42590989
|
0.971
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr11_+_117362272
|
0.953
|
NM_001558
|
IL10RA
|
interleukin 10 receptor, alpha
|
|
chrX_+_103103848
|
0.921
|
NM_194324
|
TMSB15B
|
thymosin beta 15B
|
|
chr17_+_4743645
|
0.920
|
NM_001145536
|
C17orf107
|
chromosome 17 open reading frame 107
|
|
chr19_-_55671814
|
0.913
|
NM_138411
|
FAM71E1
|
family with sequence similarity 71, member E1
|
|
chr8_+_38763911
|
0.912
|
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr8_+_79591074
|
0.890
|
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr4_+_75699652
|
0.880
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr1_-_17318452
|
0.802
|
NM_007365
|
PADI2
|
peptidyl arginine deiminase, type II
|
|
chr10_-_49483002
|
0.794
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr1_-_226712140
|
0.782
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr7_+_100111712
|
0.771
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr8_+_26204960
|
0.762
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chrX_-_151996613
|
0.735
|
NM_032882
|
PNMA6A
|
paraneoplastic antigen like 6A
|
|
chrX_+_102749489
|
0.732
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr1_+_226712430
|
0.725
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr20_+_55718619
|
0.714
|
|
|
|
|
chr22_-_42589543
|
0.709
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr18_+_55718171
|
0.706
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chrX_+_151991474
|
0.695
|
NM_001170944
NM_032882
|
LOC100287428
PNMA6A
|
paraneoplastic antigen-like protein 6A related gene
paraneoplastic antigen like 6A
|
|
chr20_+_55718718
|
0.695
|
|
|
|
|
chr5_+_177473071
|
0.694
|
NM_015111
|
N4BP3
|
NEDD4 binding protein 3
|
|
chr8_-_41641881
|
0.691
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr12_-_121946635
|
0.677
|
NM_024667
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr16_+_22733307
|
0.674
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr19_+_12917607
|
0.666
|
NM_005053
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr8_+_26204997
|
0.664
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr9_+_124066973
|
0.657
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr12_-_121946581
|
0.654
|
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae)
|
|
chr3_+_62279707
|
0.652
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chr19_+_47593137
|
0.637
|
|
|
|
|
chr14_+_104785898
|
0.625
|
NM_033271
|
BTBD6
|
BTB (POZ) domain containing 6
|
|
chr17_-_7106497
|
0.618
|
|
CLDN7
|
claudin 7
|
|
chr1_+_70443963
|
0.616
|
|
SRSF11
|
serine/arginine-rich splicing factor 11
|
|
chr5_-_1935764
|
0.613
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr14_-_87529222
|
0.613
|
|
GALC
|
galactosylceramidase
|
|
chr11_-_3775547
|
0.613
|
NM_005387
NM_016320
NM_139131
NM_139132
|
NUP98
|
nucleoporin 98kDa
|
|
chrX_-_106846684
|
0.611
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr14_+_104785936
|
0.611
|
|
BTBD6
|
BTB (POZ) domain containing 6
|
|
chr15_-_70399301
|
0.611
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr15_-_70399551
|
0.609
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr9_-_124066901
|
0.604
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr1_-_153098811
|
0.600
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chrX_-_102452461
|
0.597
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr15_-_70399523
|
0.595
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr1_-_70443851
|
0.588
|
NM_017768
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr8_-_98359248
|
0.584
|
|
TSPYL5
|
TSPY-like 5
|
|
chr15_-_70399181
|
0.581
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr14_-_87529212
|
0.572
|
|
GALC
|
galactosylceramidase
|
|
chr8_+_128875902
|
0.569
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr12_+_64869296
|
0.567
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chrX_-_106846924
|
0.566
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr7_+_20336849
|
0.560
|
|
ITGB8
|
integrin, beta 8
|
|
chr14_-_22458134
|
0.557
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr9_-_124066838
|
0.555
|
|
RBM18
|
RNA binding motif protein 18
|
|
chr12_+_64869271
|
0.550
|
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr8_-_98359347
|
0.549
|
NM_033512
|
TSPYL5
|
TSPY-like 5
|
|
chr14_-_54563554
|
0.548
|
NM_001008396
NM_007086
|
WDHD1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr12_+_64869181
|
0.546
|
NM_001142523
NM_007199
|
IRAK3
|
interleukin-1 receptor-associated kinase 3
|
|
chr8_+_26204921
|
0.537
|
NM_002717
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr19_+_12917656
|
0.530
|
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr7_-_140270749
|
0.529
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr11_+_3775529
|
0.524
|
NM_001145438
|
PGAP2
|
post-GPI attachment to proteins 2
|
|
chr11_-_3775282
|
0.523
|
|
NUP98
|
nucleoporin 98kDa
|
|
chr4_+_145786622
|
0.522
|
NM_022475
|
HHIP
|
hedgehog interacting protein
|
|
chr14_-_54563520
|
0.519
|
|
WDHD1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr3_-_121295202
|
0.517
|
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr3_-_59010797
|
0.516
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr7_-_8268206
|
0.515
|
NM_022307
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chrX_-_72351348
|
0.508
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chrX_-_72351392
|
0.508
|
NM_021963
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr3_-_59010777
|
0.499
|
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr5_-_136862317
|
0.497
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr19_+_12917671
|
0.496
|
|
RAD23A
|
RAD23 homolog A (S. cerevisiae)
|
|
chr13_-_27967215
|
0.489
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr12_-_121946561
|
0.486
|
|
|
|
|
chr8_+_56177567
|
0.486
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr4_-_113777470
|
0.485
|
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr11_-_605649
|
0.483
|
NM_004031
|
IRF7
|
interferon regulatory factor 7
|
|
chr17_+_54124928
|
0.482
|
|
RAD51C
|
RAD51 homolog C (S. cerevisiae)
|
|
chr10_+_17312654
|
0.479
|
|
VIM
|
vimentin
|
|
chr3_-_59010740
|
0.478
|
NM_198463
|
C3orf67
|
chromosome 3 open reading frame 67
|
|
chr17_+_30938391
|
0.477
|
NM_001030006
NM_001282
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr19_-_19793512
|
0.470
|
NM_001099269
NM_001145404
|
ZNF506
|
zinc finger protein 506
|
|
chr11_-_76862197
|
0.469
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chrX_+_151975421
|
0.467
|
NM_013364
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr4_-_113777569
|
0.466
|
NM_018392
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr14_-_60817621
|
0.465
|
|
|
|
|
chr12_+_4253143
|
0.465
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr7_+_116447499
|
0.465
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr1_-_153480980
|
0.460
|
NM_001005741
NM_001005742
NM_001171811
|
GBA
|
glucosidase, beta, acid
|
|
chr17_+_24095135
|
0.460
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr5_-_1547872
|
0.456
|
|
LPCAT1
|
lysophosphatidylcholine acyltransferase 1
|
|
chr3_+_130081187
|
0.455
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr7_+_100111689
|
0.453
|
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr5_-_16561936
|
0.451
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr3_+_157877146
|
0.450
|
NM_001184718
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr8_-_116750277
|
0.447
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chrX_-_13866565
|
0.444
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr14_+_54563587
|
0.444
|
NM_080867
NM_199421
|
SOCS4
|
suppressor of cytokine signaling 4
|
|
chr19_-_60587414
|
0.441
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr12_+_102883687
|
0.439
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr14_-_22458220
|
0.439
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr7_-_8268292
|
0.438
|
NM_001136020
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr1_+_91739241
|
0.435
|
NM_001134420
|
CDC7
|
cell division cycle 7 homolog (S. cerevisiae)
|
|
chr7_-_105712565
|
0.432
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr9_-_109291866
|
0.427
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr11_-_3775312
|
0.424
|
|
NUP98
|
nucleoporin 98kDa
|
|
chr14_+_23710924
|
0.422
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr17_+_45940743
|
0.422
|
NM_032133
|
MYCBPAP
|
MYCBP associated protein
|
|
chr3_+_130081161
|
0.420
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr17_+_30938433
|
0.420
|
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr4_-_84475328
|
0.420
|
NM_001166498
NM_006665
|
HPSE
|
heparanase
|
|
chr17_+_54124952
|
0.415
|
NM_002876
NM_058216
|
RAD51C
|
RAD51 homolog C (S. cerevisiae)
|
|
chr4_+_170778265
|
0.412
|
NM_001829
NM_173872
|
CLCN3
|
chloride channel 3
|
|
chr3_+_10043139
|
0.409
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr6_-_90119008
|
0.409
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr2_+_71534260
|
0.406
|
NM_001130976
NM_001130977
NM_001130978
NM_001130979
NM_001130980
NM_001130981
NM_003494
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr13_-_35818396
|
0.406
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr19_-_57082907
|
0.404
|
NM_001135590
NM_032679
|
ZNF577
|
zinc finger protein 577
|
|
chr13_-_35818491
|
0.403
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr11_-_118739838
|
0.403
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr3_+_130081149
|
0.402
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr1_+_154297588
|
0.401
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr1_+_40193368
|
0.401
|
NM_001136493
NM_032793
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr3_+_44355007
|
0.398
|
|
C3orf23
|
chromosome 3 open reading frame 23
|
|
chr2_-_166358951
|
0.398
|
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr3_+_44354927
|
0.397
|
NM_001029839
NM_173826
|
C3orf23
|
chromosome 3 open reading frame 23
|
|
chr1_+_40193424
|
0.396
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr19_-_60610792
|
0.391
|
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr15_+_76343956
|
0.388
|
NM_001130182
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr18_+_22060383
|
0.386
|
NM_005640
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa
|
|
chr1_+_40193389
|
0.384
|
|
MFSD2A
|
major facilitator superfamily domain containing 2A
|
|
chr7_+_44612745
|
0.384
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chrX_-_7855400
|
0.381
|
NM_004650
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr6_-_90119068
|
0.377
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr7_-_124357272
|
0.375
|
NM_001042594
NM_015450
|
POT1
|
protection of telomeres 1 homolog (S. pombe)
|
|
chr12_+_54838408
|
0.371
|
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr3_+_130081130
|
0.371
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr4_+_170778303
|
0.370
|
|
CLCN3
|
chloride channel 3
|
|
chr4_+_113777568
|
0.370
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr3_+_44355217
|
0.368
|
|
C3orf23
|
chromosome 3 open reading frame 23
|
|
chrX_-_72351381
|
0.367
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr7_-_150847698
|
0.362
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chrX_-_13866366
|
0.362
|
|
|
|
|
chr7_+_38184393
|
0.361
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chrX_+_151975518
|
0.358
|
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr1_+_231816372
|
0.358
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr11_+_62405736
|
0.356
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr18_+_74930379
|
0.355
|
NM_198531
|
ATP9B
|
ATPase, class II, type 9B
|
|
chr15_-_27349300
|
0.354
|
NM_138704
|
NDNL2
|
necdin-like 2
|
|
chr21_-_43671355
|
0.353
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr11_+_62405801
|
0.353
|
|
SLC3A2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2
|
|
chr6_-_90119158
|
0.353
|
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr15_-_46257611
|
0.352
|
|
MYEF2
|
myelin expression factor 2
|
|
chr1_+_70443919
|
0.349
|
NM_001190987
NM_004768
|
SRSF11
|
serine/arginine-rich splicing factor 11
|
|
chr7_-_129379947
|
0.348
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr8_-_98359366
|
0.348
|
|
TSPYL5
|
TSPY-like 5
|
|
chr19_-_55706156
|
0.348
|
NM_138334
|
JOSD2
|
Josephin domain containing 2
|
|
chr12_-_56111054
|
0.346
|
|
R3HDM2
|
R3H domain containing 2
|
|
chr1_-_199742841
|
0.345
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr6_-_90119329
|
0.342
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast)
|
|
chr1_+_208178198
|
0.341
|
|
SYT14
|
synaptotagmin XIV
|
|
chr19_-_60610971
|
0.338
|
|
UBE2S
|
ubiquitin-conjugating enzyme E2S
|
|
chr5_-_147266257
|
0.337
|
NM_206966
|
C5orf46
|
chromosome 5 open reading frame 46
|
|
chr4_-_8493327
|
0.337
|
NM_001101667
NM_003501
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr4_+_47181987
|
0.336
|
NM_020453
|
ATP10D
|
ATPase, class V, type 10D
|
|
chr1_+_89871281
|
0.332
|
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr1_+_110494654
|
0.330
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr9_-_89779180
|
0.330
|
|
CDK20
|
cyclin-dependent kinase 20
|
|
chr3_-_10042989
|
0.329
|
|
CIDECP
|
cell death-inducing DFFA-like effector c pseudogene
|
|
chr12_+_54838361
|
0.326
|
NM_021019
NM_079423
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|
|
chr1_-_208024491
|
0.324
|
NM_152485
|
C1orf74
|
chromosome 1 open reading frame 74
|