|
chr13_+_110653407
|
2.594
|
|
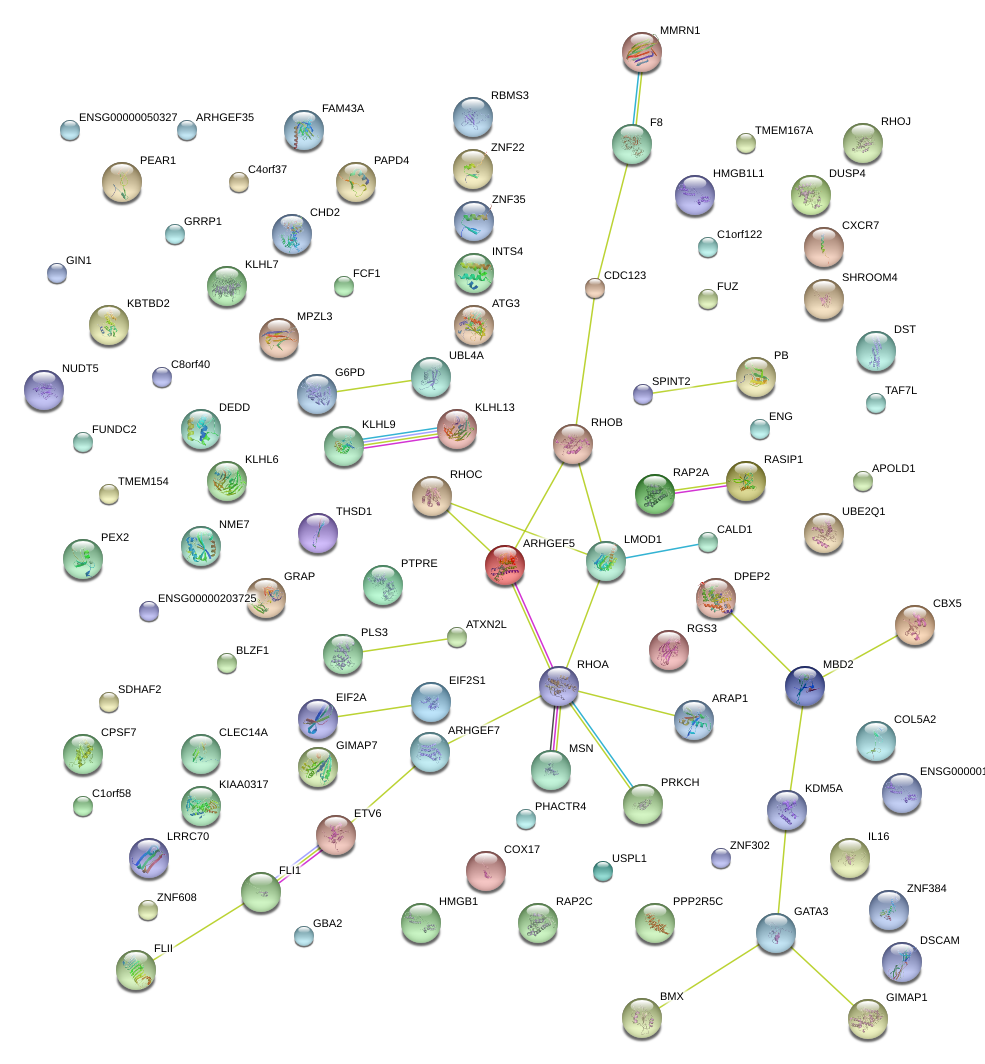
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr13_-_30089509
|
2.250
|
|
HMGB1
|
high-mobility group box 1
|
|
chr14_+_62740878
|
2.244
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr19_+_43447262
|
2.103
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr7_+_150044530
|
2.092
|
NM_130759
|
GIMAP1
|
GTPase, IMAP family member 1
|
|
chr5_+_61910318
|
2.031
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr5_-_124108672
|
1.993
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr3_-_184756101
|
1.880
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr4_+_91035025
|
1.842
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chrX_+_114734074
|
1.807
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr11_-_72111027
|
1.679
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr9_+_115303527
|
1.542
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr10_+_8137356
|
1.512
|
|
GATA3
|
GATA binding protein 3
|
|
chr1_+_26358097
|
1.496
|
NM_024869
|
GRRP1
|
glycine/arginine rich protein 1
|
|
chr7_-_143622188
|
1.478
|
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr1_+_167604056
|
1.459
|
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr3_+_29297806
|
1.458
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr17_-_74244539
|
1.411
|
|
|
|
|
chr22_-_34566210
|
1.404
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr3_-_120878910
|
1.403
|
NM_005694
|
COX17
|
COX17 cytochrome c oxidase assembly homolog (S. cerevisiae)
|
|
chr17_-_18890963
|
1.386
|
NM_006613
|
GRAP
|
GRB2-related adaptor protein
|
|
chr7_-_143523582
|
1.384
|
NM_001003702
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chr8_-_29262240
|
1.370
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr7_-_143622233
|
1.345
|
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chrX_-_50403412
|
1.344
|
|
SHROOM4
|
shroom family member 4
|
|
chr7_+_149842850
|
1.318
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr22_-_34566276
|
1.306
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr13_+_30089829
|
1.304
|
NM_005800
|
USPL1
|
ubiquitin specific peptidase like 1
|
|
chr7_+_143683365
|
1.301
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr1_+_28637247
|
1.296
|
NM_023923
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr14_-_37795001
|
1.269
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr3_-_113763049
|
1.266
|
NM_022488
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chr12_-_52939636
|
1.232
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr2_+_237143118
|
1.230
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr22_-_34566367
|
1.219
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr3_+_195889845
|
1.204
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr12_-_368517
|
1.185
|
|
KDM5A
|
lysine (K)-specific demethylase 5A
|
|
chr10_+_12278213
|
1.180
|
|
CDC123
|
cell division cycle 123 homolog (S. cerevisiae)
|
|
chr3_+_44665220
|
1.176
|
NM_003420
|
ZNF35
|
zinc finger protein 35
|
|
chr1_+_167603817
|
1.171
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chr14_-_74249492
|
1.168
|
NM_001039479
|
KIAA0317
|
KIAA0317
|
|
chr10_-_12278026
|
1.165
|
NM_014142
|
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5
|
|
chr5_+_78944151
|
1.153
|
|
PAPD4
|
PAP associated domain containing 4
|
|
chr1_-_152797707
|
1.145
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr10_-_112668679
|
1.140
|
NM_001195306
|
BBIP1
|
BBSome interacting protein 1
|
|
chr8_-_78074851
|
1.139
|
NM_000318
NM_001079867
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr12_+_11694172
|
1.133
|
|
ETV6
|
ets variant 6
|
|
chr14_-_37795290
|
1.132
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr19_+_43447078
|
1.130
|
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr1_-_152797760
|
1.119
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr6_-_76050837
|
1.114
|
|
|
|
|
chr1_+_150428404
|
1.113
|
|
|
|
|
chr19_+_39860398
|
1.103
|
NM_001012320
NM_018443
|
ZNF302
|
zinc finger protein 302
|
|
chr10_+_129735796
|
1.102
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr19_-_53935648
|
1.102
|
NM_017805
|
RASIP1
|
Ras interacting protein 1
|
|
chr6_-_56824672
|
1.099
|
|
DST
|
dystonin
|
|
chr11_-_117628211
|
1.091
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr15_+_91248706
|
1.089
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr2_+_24152799
|
1.089
|
|
|
|
|
chrX_-_153428621
|
1.087
|
|
G6PD
|
glucose-6-phosphate dehydrogenase
|
|
chr11_-_60953848
|
1.085
|
NM_024811
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa
|
|
chr21_+_25856311
|
1.084
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr9_-_35738875
|
1.078
|
|
GBA2
|
glucosidase, beta (bile acid) 2
|
|
chr14_+_74249599
|
1.075
|
NM_015962
|
FCF1
|
FCF1 small subunit (SSU) processome component homolog (S. cerevisiae)
|
|
chr10_-_112668892
|
1.073
|
NM_001195304
NM_001195305
NM_001195307
|
BBIP1
|
BBSome interacting protein 1
|
|
chr10_+_12278185
|
1.070
|
|
CDC123
|
cell division cycle 123 homolog (S. cerevisiae)
|
|
chr4_-_153820535
|
1.064
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr1_-_159369081
|
1.058
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr15_+_79376295
|
1.039
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr1_-_167603623
|
1.034
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr12_-_6668880
|
1.029
|
NM_001039917
NM_001039919
NM_001039920
NM_001135734
|
ZNF384
|
zinc finger protein 384
|
|
chr1_+_220953278
|
1.027
|
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr4_-_99283323
|
1.025
|
NM_174952
|
C4orf37
|
chromosome 4 open reading frame 37
|
|
chr11_+_60954096
|
1.023
|
|
SDHAF2
|
succinate dehydrogenase complex assembly factor 2
|
|
chr7_-_32897714
|
1.017
|
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr10_+_12278200
|
1.015
|
|
CDC123
|
cell division cycle 123 homolog (S. cerevisiae)
|
|
chr3_-_49424322
|
1.011
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr1_-_159369041
|
1.008
|
|
DEDD
|
death effector domain containing
|
|
chr5_-_102483680
|
1.006
|
NM_017676
|
GIN1
|
gypsy retrotransposon integrase 1
|
|
chr1_+_220953351
|
1.005
|
|
BROX
|
BRO1 domain and CAAX motif containing
|
|
chr14_+_101346032
|
0.995
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr11_+_128069165
|
0.989
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr1_+_38046404
|
0.987
|
|
C1orf122
|
chromosome 1 open reading frame 122
|
|
chr14_-_56805118
|
0.984
|
|
|
|
|
chrX_-_153908408
|
0.981
|
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr14_+_60858151
|
0.978
|
NM_006255
|
PRKCH
|
protein kinase C, eta
|
|
chrX_-_131179598
|
0.976
|
NM_021183
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr9_-_21325239
|
0.975
|
NM_018847
|
KLHL9
|
kelch-like 9 (Drosophila)
|
|
chr11_+_128069183
|
0.969
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr1_-_152797767
|
0.965
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr1_-_152797820
|
0.964
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr13_-_51878235
|
0.964
|
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr18_-_50004770
|
0.963
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr19_-_55008172
|
0.963
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr5_-_82408917
|
0.962
|
|
TMEM167A
|
transmembrane protein 167A
|
|
chr11_+_65027721
|
0.959
|
|
|
|
|
chr11_-_77383359
|
0.950
|
NM_033547
|
INTS4
|
integrator complex subunit 4
|
|
chrX_+_64804318
|
0.946
|
|
MSN
|
moesin
|
|
chrX_+_64804235
|
0.945
|
NM_002444
|
MSN
|
moesin
|
|
chr2_-_189752575
|
0.945
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr3_-_49424361
|
0.940
|
|
RHOA
|
ras homolog gene family, member A
|
|
chrX_+_64804259
|
0.936
|
|
MSN
|
moesin
|
|
chrX_+_15428820
|
0.935
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr17_-_60208083
|
0.932
|
|
LOC146880
|
hypothetical LOC146880
|
|
chr7_+_134114701
|
0.931
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr1_+_155130146
|
0.931
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr7_+_134114912
|
0.929
|
|
CALD1
|
caldesmon 1
|
|
chr19_+_43446937
|
0.928
|
NM_001166103
NM_021102
|
SPINT2
|
serine peptidase inhibitor, Kunitz type, 2
|
|
chr3_+_151747204
|
0.928
|
NM_032025
|
EIF2A
|
eukaryotic translation initiation factor 2A, 65kDa
|
|
chr8_+_42515873
|
0.925
|
NM_138436
NM_001135674
NM_001135675
|
C8orf40
|
chromosome 8 open reading frame 40
|
|
chr11_+_128069203
|
0.925
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr11_-_60954031
|
0.924
|
NM_001136040
NM_001142565
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa
|
|
chrX_+_153908426
|
0.922
|
|
FUNDC2
|
FUN14 domain containing 2
|
|
chr11_+_60954150
|
0.917
|
NM_017841
|
SDHAF2
|
succinate dehydrogenase complex assembly factor 2
|
|
chr10_+_44816278
|
0.916
|
NM_006963
|
ZNF22
|
zinc finger protein 22 (KOX 15)
|
|
chr11_-_77383307
|
0.916
|
|
INTS4
|
integrator complex subunit 4
|
|
chr9_-_129656581
|
0.915
|
|
ENG
|
endoglin
|
|
chr5_+_78944052
|
0.914
|
|
PAPD4
|
PAP associated domain containing 4
|
|
chr12_+_12829807
|
0.912
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr19_+_3129735
|
0.907
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr15_-_90999606
|
0.907
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr6_-_42293577
|
0.906
|
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr5_-_82409009
|
0.903
|
NM_174909
|
TMEM167A
|
transmembrane protein 167A
|
|
chr3_+_185515543
|
0.902
|
NM_182917
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr9_-_129656413
|
0.900
|
|
ENG
|
endoglin
|
|
chr6_-_42293581
|
0.899
|
NM_018141
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr3_-_113763217
|
0.893
|
|
ATG3
|
ATG3 autophagy related 3 homolog (S. cerevisiae)
|
|
chrX_-_40479607
|
0.893
|
|
MED14
|
mediator complex subunit 14
|
|
chr11_+_128068982
|
0.891
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr19_-_42098919
|
0.883
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr8_-_11096257
|
0.878
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr12_-_74191649
|
0.876
|
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr3_+_9809776
|
0.876
|
NM_001198780
|
ARPC4
|
actin related protein 2/3 complex, subunit 4, 20kDa
|
|
chr19_+_44266784
|
0.876
|
NM_001004318
|
PAPL
|
iron/zinc purple acid phosphatase-like protein
|
|
chr8_-_103735194
|
0.874
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr1_+_84382575
|
0.870
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr10_+_12277935
|
0.866
|
NM_006023
|
CDC123
|
cell division cycle 123 homolog (S. cerevisiae)
|
|
chr12_+_368774
|
0.864
|
NM_001130147
NM_001130148
|
CCDC77
|
coiled-coil domain containing 77
|
|
chrX_-_40479893
|
0.864
|
|
MED14
|
mediator complex subunit 14
|
|
chr12_-_74191607
|
0.857
|
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr5_-_112658200
|
0.856
|
|
MCC
|
mutated in colorectal cancers
|
|
chr12_-_74191658
|
0.853
|
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr8_-_59734474
|
0.853
|
NM_001144772
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor
|
|
chr3_-_49424404
|
0.852
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr13_-_44813203
|
0.852
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chrX_+_153428755
|
0.850
|
NM_001099857
NM_001145255
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr2_-_24152813
|
0.849
|
NM_016047
|
SF3B14
|
splicing factor 3B, 14 kDa subunit
|
|
chr5_+_78943998
|
0.846
|
NM_001114393
NM_001114394
NM_173797
|
PAPD4
|
PAP associated domain containing 4
|
|
chr7_+_134114957
|
0.845
|
|
CALD1
|
caldesmon 1
|
|
chr9_+_99435900
|
0.844
|
|
NCBP1
|
nuclear cap binding protein subunit 1, 80kDa
|
|
chr5_+_82409072
|
0.843
|
NM_003401
NM_022406
NM_022550
|
XRCC4
|
X-ray repair complementing defective repair in Chinese hamster cells 4
|
|
chr14_+_101345878
|
0.842
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr1_-_38046270
|
0.833
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr3_-_49424380
|
0.833
|
|
RHOA
|
ras homolog gene family, member A
|
|
chr11_+_128069340
|
0.831
|
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr5_+_102483856
|
0.830
|
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr10_-_103567773
|
0.830
|
|
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase)
|
|
chr1_+_84382562
|
0.827
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr16_-_21739231
|
0.825
|
|
RRN3P1
|
RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene 1
|
|
chrX_+_153908257
|
0.819
|
NM_023934
|
FUNDC2
|
FUN14 domain containing 2
|
|
chr16_-_3567354
|
0.819
|
NM_178844
|
NLRC3
|
NLR family, CARD domain containing 3
|
|
chr11_-_85457469
|
0.819
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr12_-_74191679
|
0.816
|
NM_007043
|
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast)
|
|
chr5_-_151046638
|
0.815
|
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr2_+_33514896
|
0.814
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr12_-_52939584
|
0.812
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr9_-_129656778
|
0.810
|
|
ENG
|
endoglin
|
|
chr7_+_44207163
|
0.809
|
|
YKT6
|
YKT6 v-SNARE homolog (S. cerevisiae)
|
|
chr6_-_42293564
|
0.806
|
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr1_+_39229474
|
0.805
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr3_-_49424397
|
0.799
|
|
RHOA
|
ras homolog gene family, member A
|
|
chrX_+_24621984
|
0.799
|
NM_016937
|
POLA1
|
polymerase (DNA directed), alpha 1, catalytic subunit
|
|
chr1_-_38046433
|
0.798
|
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chr6_-_42293575
|
0.797
|
|
MRPS10
|
mitochondrial ribosomal protein S10
|
|
chr15_-_40352765
|
0.796
|
NM_001110503
NM_015497
|
TMEM87A
|
transmembrane protein 87A
|
|
chr4_-_90978469
|
0.795
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chrX_+_119622570
|
0.793
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr1_-_38046439
|
0.792
|
NM_024640
|
YRDC
|
yrdC domain containing (E. coli)
|
|
chrX_+_51562894
|
0.792
|
NM_001005332
|
MAGED1
|
melanoma antigen family D, 1
|
|
chr3_-_195874227
|
0.789
|
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chr12_+_120840829
|
0.788
|
NM_001178003
NM_144668
|
WDR66
|
WD repeat domain 66
|
|
chr22_-_17892859
|
0.788
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr3_-_195874170
|
0.787
|
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chr19_-_39859997
|
0.785
|
|
|
|
|
chr11_+_73559900
|
0.784
|
|
PPME1
|
protein phosphatase methylesterase 1
|
|
chr14_+_77244177
|
0.781
|
NM_031210
|
C14orf156
|
chromosome 14 open reading frame 156
|
|
chr3_-_195874202
|
0.779
|
|
LSG1
|
large subunit GTPase 1 homolog (S. cerevisiae)
|
|
chr14_-_77244073
|
0.776
|
NM_006020
|
ALKBH1
|
alkB, alkylation repair homolog 1 (E. coli)
|
|
chr10_+_75174151
|
0.775
|
|
SEC24C
|
SEC24 family, member C (S. cerevisiae)
|
|
chrX_+_77052834
|
0.774
|
NM_000052
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr1_-_40496161
|
0.773
|
|
|
|
|
chr5_-_151046680
|
0.773
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr3_-_71262405
|
0.773
|
|
FOXP1
|
forkhead box P1
|
|
chr20_+_3818441
|
0.771
|
NM_153640
|
PANK2
|
pantothenate kinase 2
|
|
chr14_-_22458134
|
0.770
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr1_+_84382529
|
0.766
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr7_-_91347915
|
0.766
|
NM_006980
|
MTERF
|
mitochondrial transcription termination factor
|
|
chr5_-_102926349
|
0.764
|
NM_031438
|
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12
|
|
chr10_+_75174125
|
0.762
|
NM_004922
NM_198597
|
SEC24C
|
SEC24 family, member C (S. cerevisiae)
|
|
chr17_+_4922478
|
0.761
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|