|
chr1_+_156229934
|
5.106
|
|
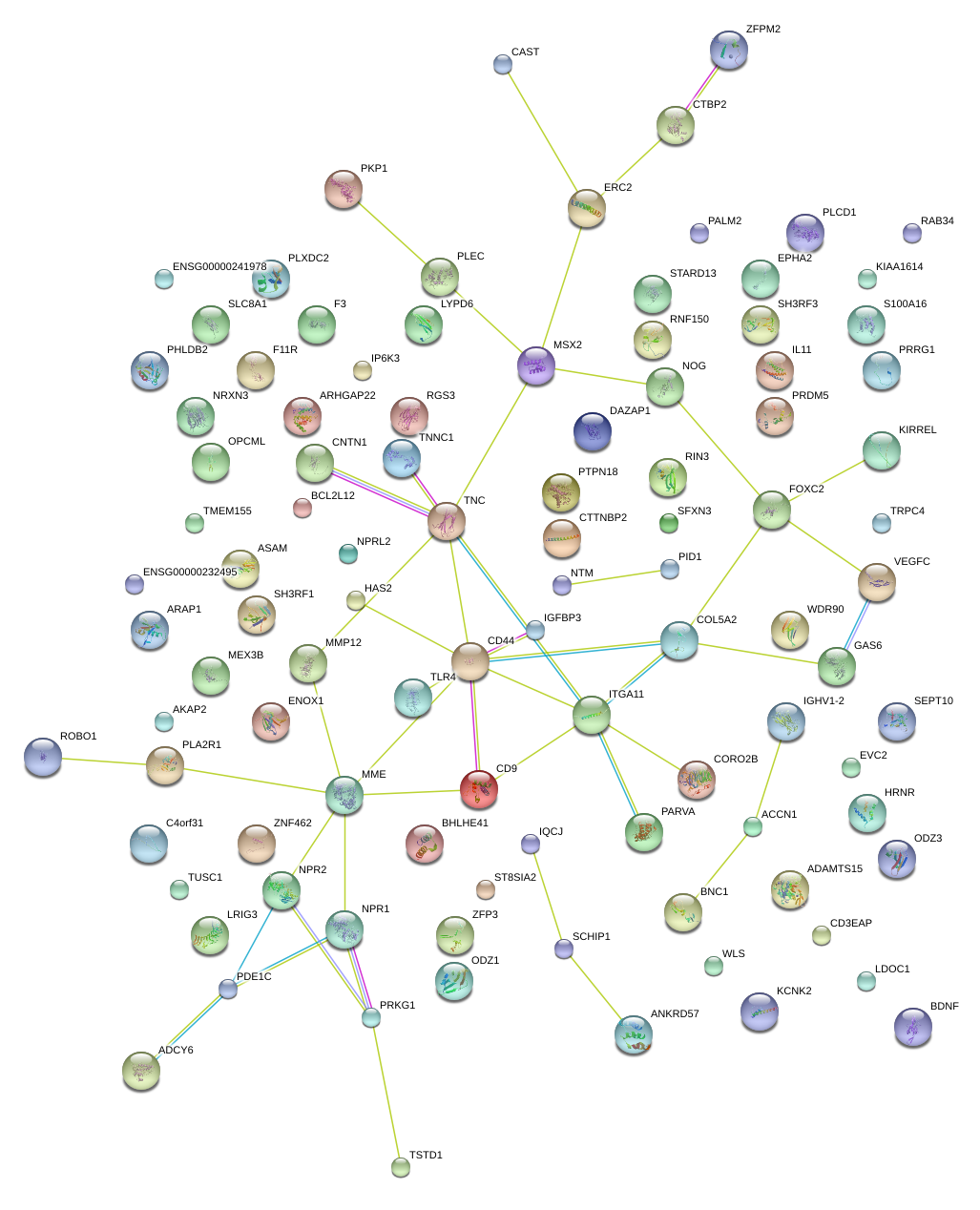
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr10_-_126839546
|
4.692
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr5_+_96024434
|
4.552
|
|
CAST
|
calpastatin
|
|
chr5_+_174084137
|
4.357
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr12_+_6179736
|
3.679
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr8_-_122722674
|
3.655
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr4_+_183302105
|
3.435
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr8_-_145085616
|
3.345
|
NM_201384
|
PLEC
|
plectin
|
|
chr13_-_43259032
|
3.212
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr2_+_109729176
|
3.188
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr3_+_160964574
|
3.159
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_-_26169074
|
3.122
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr3_-_38046034
|
3.101
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr10_-_49483002
|
3.050
|
NM_021226
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr11_+_131286513
|
3.037
|
|
NTM
|
neurotrimin
|
|
chr9_+_35782405
|
2.961
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr15_-_66511488
|
2.952
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr3_+_156280539
|
2.940
|
|
MME
|
membrane metallo-endopeptidase
|
|
chr4_-_122213018
|
2.884
|
NM_024574
|
C4orf31
|
chromosome 4 open reading frame 31
|
|
chr9_-_116920232
|
2.734
|
NM_002160
|
TNC
|
tenascin C
|
|
chr3_+_156280497
|
2.734
|
|
MME
|
membrane metallo-endopeptidase
|
|
chr1_-_151852128
|
2.685
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr1_+_179148935
|
2.677
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr17_-_24068759
|
2.660
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr4_-_177950658
|
2.639
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr15_+_66658626
|
2.614
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr1_+_156229686
|
2.609
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr9_+_111442888
|
2.604
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chrX_-_140098941
|
2.578
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr6_-_72187168
|
2.569
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr12_-_47468985
|
2.555
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr9_+_108665168
|
2.537
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr1_+_213323182
|
2.497
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr2_+_130846263
|
2.458
|
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chr2_-_109728958
|
2.435
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr3_+_113061330
|
2.406
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr8_+_106400322
|
2.391
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr15_-_81743198
|
2.377
|
|
BNC1
|
basonuclin 1
|
|
chr3_+_113061296
|
2.365
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr17_-_76985538
|
2.315
|
|
|
|
|
chr7_-_45927263
|
2.311
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr1_-_159275357
|
2.279
|
NM_001113205
NM_001113206
NM_001113207
|
TSTD1
F11R
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
F11 receptor
|
|
chrX_-_140098871
|
2.277
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chrX_-_140098959
|
2.265
|
NM_012317
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr3_+_113061218
|
2.236
|
NM_001134439
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr2_-_40532651
|
2.185
|
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chrX_-_140099014
|
2.178
|
|
LDOC1
|
leucine zipper, down-regulated in cancer 1
|
|
chr1_-_16355013
|
2.151
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr15_+_90738108
|
2.135
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr2_-_189752575
|
2.108
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr2_-_56266408
|
2.078
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr16_+_85158357
|
2.060
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr1_-_94779727
|
2.056
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr3_-_79151232
|
2.053
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr4_-_142273881
|
2.047
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr9_-_25668230
|
2.027
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr13_-_32822759
|
2.020
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chrX_+_37093448
|
2.008
|
NM_000950
NM_001142395
NM_001173486
NM_001173489
NM_001173490
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1
|
|
chr11_+_35117226
|
1.990
|
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr10_+_102782156
|
1.987
|
|
SFXN3
|
sideroflexin 3
|
|
chr7_-_117300763
|
1.958
|
NM_033427
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr10_+_52504239
|
1.945
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr13_-_37341859
|
1.933
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr1_+_199519202
|
1.932
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr14_+_78815406
|
1.927
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr2_-_160627366
|
1.901
|
NM_001007267
NM_001195641
NM_007366
|
PLA2R1
|
phospholipase A2 receptor 1, 180kDa
|
|
chr11_-_122571198
|
1.896
|
NM_024769
|
CLMP
|
CXADR-like membrane protein
|
|
chr2_+_109112428
|
1.891
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr7_-_32076985
|
1.876
|
NM_001191056
NM_001191057
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa
|
|
chr13_+_113546841
|
1.860
|
NM_000820
|
GAS6
|
growth arrest-specific 6
|
|
chr3_+_156280604
|
1.857
|
NM_007288
|
MME
|
membrane metallo-endopeptidase
|
|
chr17_+_4922445
|
1.851
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr4_-_187884721
|
1.849
|
|
|
|
|
chr2_+_149895274
|
1.849
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr11_-_72111027
|
1.836
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr1_-_68470585
|
1.835
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr4_-_122063118
|
1.832
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr10_+_20145350
|
1.825
|
NM_032812
|
PLXDC2
|
plexin domain containing 2
|
|
chr9_+_119506428
|
1.817
|
|
TLR4
|
toll-like receptor 4
|
|
chr4_-_5761194
|
1.817
|
NM_147127
|
EVC2
|
Ellis van Creveld syndrome 2
|
|
chr19_-_60573552
|
1.812
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr3_+_156280129
|
1.801
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr9_+_115396020
|
1.793
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr2_-_229844198
|
1.792
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr2_-_160627181
|
1.790
|
|
PLA2R1
|
phospholipase A2 receptor 1, 180kDa
|
|
chr17_+_4922478
|
1.784
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr17_+_52026058
|
1.763
|
NM_005450
|
NOG
|
noggin
|
|
chr15_-_80123935
|
1.734
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr16_+_639278
|
1.695
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr4_-_122905731
|
1.686
|
NM_152399
|
TMEM155
|
transmembrane protein 155
|
|
chr14_+_92049396
|
1.677
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr11_-_27677755
|
1.672
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_+_12355600
|
1.667
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr11_+_129823667
|
1.632
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr12_-_57600564
|
1.627
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr6_-_33822659
|
1.620
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr12_+_52653176
|
1.619
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr18_-_20231783
|
1.607
|
NM_080597
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr7_-_32077504
|
1.601
|
NM_001191059
NM_005020
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa
|
|
chr11_+_696607
|
1.597
|
|
EPS8L2
|
EPS8-like 2
|
|
chr10_+_46413551
|
1.593
|
NM_014696
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2
|
|
chr9_+_126671321
|
1.592
|
|
ARPC5L
|
actin related protein 2/3 complex, subunit 5-like
|
|
chr2_+_182030173
|
1.589
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr10_+_52421120
|
1.583
|
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr18_-_20231709
|
1.578
|
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr4_+_167013859
|
1.575
|
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr1_+_203064397
|
1.573
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr13_-_39075304
|
1.567
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chrX_-_1470862
|
1.562
|
NM_001636
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6
|
|
chr19_+_46417117
|
1.556
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr5_+_178300799
|
1.549
|
NM_001178089
NM_001178090
NM_182594
|
ZNF454
|
zinc finger protein 454
|
|
chr12_+_52806103
|
1.548
|
|
LOC400043
|
hypothetical LOC400043
|
|
chr11_+_104986620
|
1.535
|
NM_001112812
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr9_-_20612433
|
1.528
|
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr19_+_10900502
|
1.527
|
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr2_+_191818124
|
1.523
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr11_+_124539814
|
1.519
|
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr17_-_72045218
|
1.517
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr10_-_116154504
|
1.512
|
NM_001001936
NM_032550
|
AFAP1L2
|
actin filament associated protein 1-like 2
|
|
chr9_+_71848555
|
1.510
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr10_+_52420936
|
1.505
|
NM_001098512
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr11_+_92342436
|
1.505
|
NM_005959
|
MTNR1B
|
melatonin receptor 1B
|
|
chr13_+_113546897
|
1.486
|
|
GAS6
|
growth arrest-specific 6
|
|
chrX_-_99551926
|
1.483
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr11_+_35116961
|
1.483
|
NM_000610
NM_001001389
NM_001001390
NM_001001391
NM_001001392
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr5_+_31229549
|
1.482
|
NM_004932
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr10_-_30388439
|
1.472
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr19_-_61680510
|
1.472
|
NM_022103
|
ZNF667
|
zinc finger protein 667
|
|
chr3_+_156280338
|
1.470
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr22_+_40800213
|
1.466
|
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr13_+_113546912
|
1.464
|
|
GAS6
|
growth arrest-specific 6
|
|
chr17_+_76988134
|
1.457
|
NM_001080519
|
BAHCC1
|
BAH domain and coiled-coil containing 1
|
|
chr1_+_202308814
|
1.457
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr2_-_109728693
|
1.449
|
|
SEPT10
|
septin 10
|
|
chr9_+_119506446
|
1.429
|
|
TLR4
|
toll-like receptor 4
|
|
chr4_-_186693574
|
1.428
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr5_-_92982927
|
1.428
|
|
|
|
|
chr16_-_85099941
|
1.425
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr7_+_128257698
|
1.425
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr18_-_33399997
|
1.424
|
NM_001025087
NM_001025088
NM_001025089
NM_020180
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr6_-_112682459
|
1.415
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr2_+_182029863
|
1.405
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr11_+_24475091
|
1.400
|
NM_001009909
|
LUZP2
|
leucine zipper protein 2
|
|
chr15_+_68971835
|
1.391
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr3_-_31998241
|
1.390
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr10_+_71232281
|
1.386
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr13_+_113546943
|
1.384
|
|
GAS6
|
growth arrest-specific 6
|
|
chr2_+_54537842
|
1.379
|
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr1_+_56883333
|
1.379
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr4_+_61749540
|
1.375
|
|
LPHN3
|
latrophilin 3
|
|
chr22_+_40800193
|
1.373
|
NM_001002034
|
FAM109B
|
family with sequence similarity 109, member B
|
|
chr5_-_83716346
|
1.361
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr3_+_44729138
|
1.360
|
NM_001134440
NM_001134441
NM_001134442
NM_033210
|
ZNF502
|
zinc finger protein 502
|
|
chr8_-_23077392
|
1.352
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr19_+_10900419
|
1.351
|
NM_138358
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr6_+_43151922
|
1.351
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr12_-_57600336
|
1.349
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr20_+_60810610
|
1.329
|
NM_002531
|
NTSR1
|
neurotensin receptor 1 (high affinity)
|
|
chr2_-_225615556
|
1.324
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr7_-_107883929
|
1.324
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr11_+_128266460
|
1.316
|
NM_000890
|
KCNJ5
|
potassium inwardly-rectifying channel, subfamily J, member 5
|
|
chr3_+_112273352
|
1.314
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr17_+_58058783
|
1.311
|
|
MRC2
|
mannose receptor, C type 2
|
|
chr4_-_114901672
|
1.310
|
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta
|
|
chr19_+_61711008
|
1.309
|
NM_020813
|
ZNF471
|
zinc finger protein 471
|
|
chr9_-_128924706
|
1.307
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr3_+_157875408
|
1.303
|
NM_001184717
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr19_-_51608665
|
1.296
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr15_-_21483457
|
1.295
|
|
NDN
|
necdin homolog (mouse)
|
|
chr9_-_25668855
|
1.293
|
NM_001004125
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr11_+_124539768
|
1.292
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr5_-_92942680
|
1.282
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr7_+_55054152
|
1.278
|
NM_005228
NM_201282
NM_201283
NM_201284
|
EGFR
|
epidermal growth factor receptor
|
|
chr11_-_27700180
|
1.261
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr12_-_47605478
|
1.254
|
NM_001143782
NM_016594
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr10_+_53744043
|
1.249
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chrX_-_54538591
|
1.241
|
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr19_+_61681294
|
1.234
|
|
LOC100128252
|
hypothetical LOC100128252
|
|
chr10_+_88718167
|
1.231
|
NM_006829
|
C10orf116
|
chromosome 10 open reading frame 116
|
|
chr16_-_52877868
|
1.231
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr9_-_128924782
|
1.229
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr19_+_61681361
|
1.228
|
|
LOC100128252
|
hypothetical LOC100128252
|
|
chr3_+_48482625
|
1.228
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr10_+_88720477
|
1.224
|
NM_133447
|
AGAP11
|
ankyrin repeat and GTPase domain Arf GTPase activating protein 11
|
|
chr5_-_33927724
|
1.224
|
NM_030955
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12
|
|
chr15_-_21483503
|
1.223
|
NM_002487
|
NDN
|
necdin homolog (mouse)
|
|
chr20_-_4177519
|
1.223
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr3_+_113060716
|
1.220
|
NM_001134438
NM_145753
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr18_-_55515623
|
1.214
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr4_+_156900053
|
1.214
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chrX_-_3274680
|
1.214
|
NM_015419
|
MXRA5
|
matrix-remodelling associated 5
|
|
chr11_+_46678892
|
1.210
|
NM_024741
|
ZNF408
|
zinc finger protein 408
|
|
chr3_+_113060826
|
1.210
|
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr9_+_119506274
|
1.208
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr5_-_146869613
|
1.203
|
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr14_+_66044958
|
1.194
|
|
GPHN
|
gephyrin
|
|
chr14_+_55654900
|
1.191
|
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr10_-_131652007
|
1.187
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr1_+_182622850
|
1.184
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr9_+_33740463
|
1.179
|
NM_001197097
NM_007343
NM_001197098
|
PRSS3
|
protease, serine, 3
|