|
chr3_-_122862391
|
5.778
|
|
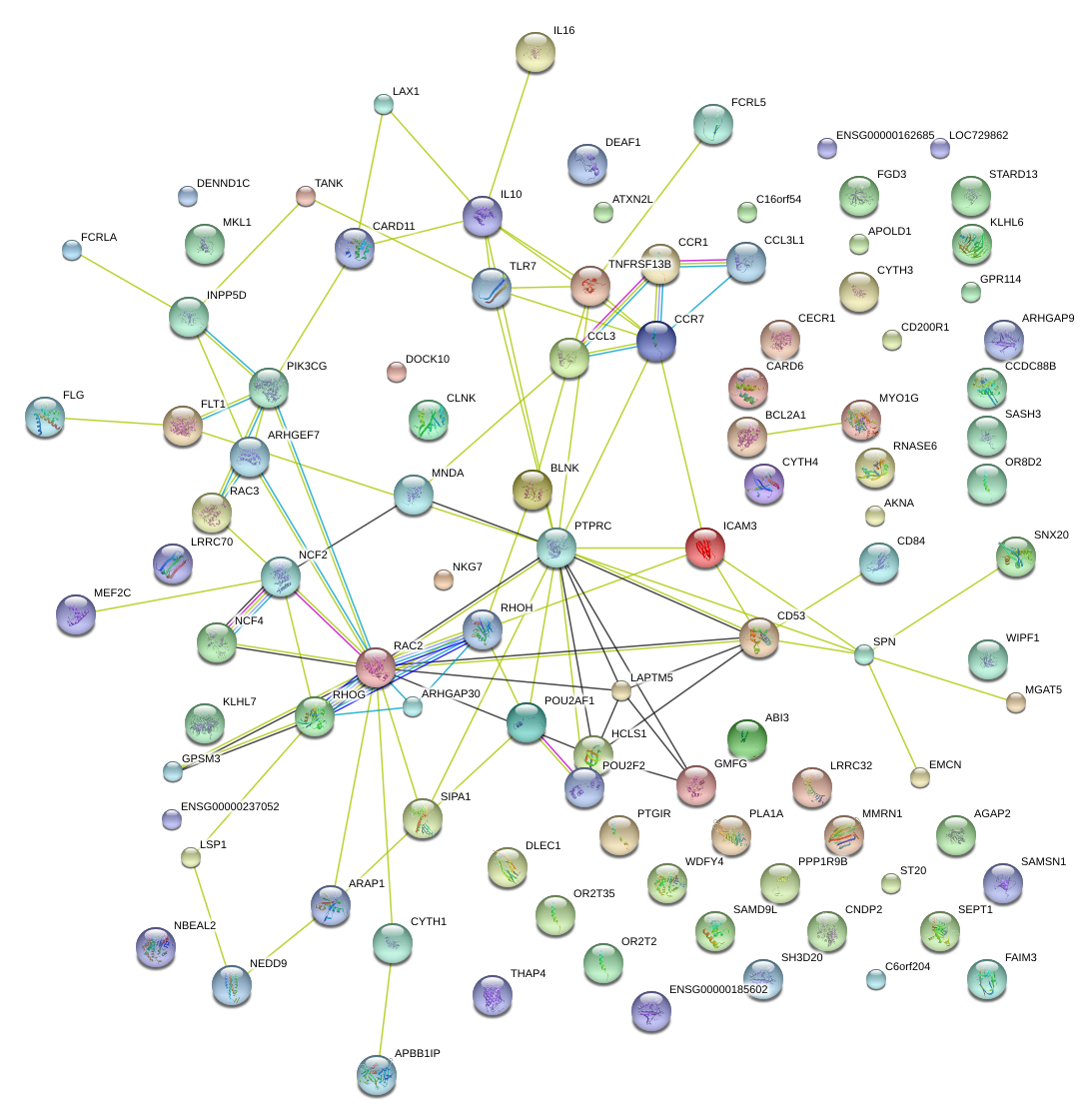
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr3_-_184756101
|
5.702
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr3_-_122862447
|
5.685
|
NM_005335
|
HCLS1
|
hematopoietic cell-specific Lyn substrate 1
|
|
chr7_+_106292975
|
5.066
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr12_-_56157898
|
5.015
|
NM_001080156
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr15_+_79376295
|
4.965
|
NM_004513
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr4_+_39874921
|
4.962
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr7_+_106292932
|
4.937
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr2_-_175207521
|
4.661
|
NM_003387
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr11_+_65164167
|
4.636
|
NM_153253
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr5_+_61910318
|
4.532
|
NM_181506
|
LRRC70
|
leucine rich repeat containing 70
|
|
chr17_-_31441390
|
4.307
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr10_+_49562866
|
4.257
|
|
WDFY4
|
WDFY family member 4
|
|
chr13_+_110653407
|
4.092
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr10_-_98021144
|
4.072
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr2_+_161701711
|
3.954
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr21_-_14840506
|
3.904
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr3_-_46224791
|
3.833
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr17_-_40843530
|
3.658
|
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr6_-_32268267
|
3.605
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr17_-_16816113
|
3.604
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr1_-_155788775
|
3.593
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr11_-_72111027
|
3.551
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr7_+_106293112
|
3.541
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr3_-_13032130
|
3.529
|
|
|
|
|
chr19_-_44518465
|
3.438
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr1_-_205161821
|
3.429
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr3_-_114176401
|
3.396
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr5_-_88214720
|
3.365
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_+_134728299
|
3.354
|
NM_002410
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase
|
|
chr14_+_20319049
|
3.351
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr6_-_11490487
|
3.342
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr1_+_111217244
|
3.339
|
NM_000560
|
CD53
|
CD53 molecule
|
|
chr1_-_205012348
|
3.290
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr19_-_10311262
|
3.273
|
NM_002162
|
ICAM3
|
intercellular adhesion molecule 3
|
|
chr22_-_39189369
|
3.190
|
|
MKL1
|
megakaryoblastic leukemia (translocation) 1
|
|
chr1_+_111217314
|
3.170
|
|
CD53
|
CD53 molecule
|
|
chr4_+_91035025
|
3.129
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr13_-_27967215
|
3.103
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr1_-_111544603
|
3.063
|
|
|
|
|
chr7_-_3049872
|
3.013
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr2_-_225519974
|
3.004
|
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr1_+_111544469
|
2.962
|
|
|
|
|
chr4_-_10295412
|
2.937
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chr22_+_35586950
|
2.927
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr10_+_26767657
|
2.887
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr5_+_40877053
|
2.876
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr16_-_29664765
|
2.865
|
NM_175900
|
C16orf54
|
chromosome 16 open reading frame 54
|
|
chr6_-_32268615
|
2.856
|
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr19_-_56567602
|
2.855
|
NM_005601
|
NKG7
|
natural killer cell group 7 sequence
|
|
chr11_+_65164767
|
2.827
|
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr19_-_6432765
|
2.817
|
NM_024898
|
DENND1C
|
DENN/MADD domain containing 1C
|
|
chr9_-_116190038
|
2.808
|
|
AKNA
|
AT-hook transcription factor
|
|
chr1_+_202000906
|
2.800
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr19_-_47328399
|
2.797
|
NM_002698
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr10_+_26767698
|
2.714
|
|
APBB1IP
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein
|
|
chr1_-_246869181
|
2.711
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr17_-_35975227
|
2.707
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr10_+_49563520
|
2.639
|
NM_020945
|
WDFY4
|
WDFY family member 4
|
|
chr1_+_159943385
|
2.637
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chrX_+_12795111
|
2.611
|
NM_016562
|
TLR7
|
toll-like receptor 7
|
|
chr1_-_181826086
|
2.582
|
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr3_+_120799411
|
2.582
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr19_-_44518516
|
2.580
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr1_-_159306037
|
2.556
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr1_-_181826292
|
2.544
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr6_-_119137923
|
2.535
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr11_+_63864263
|
2.505
|
NM_032251
|
CCDC88B
|
coiled-coil domain containing 88B
|
|
chr4_-_101658094
|
2.497
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr16_+_56134037
|
2.473
|
|
GPR114
|
G protein-coupled receptor 114
|
|
chr16_+_56134077
|
2.467
|
NM_153837
|
GPR114
|
G protein-coupled receptor 114
|
|
chr7_-_44985175
|
2.458
|
|
MYO1G
|
myosin IG
|
|
chr9_+_94766379
|
2.447
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr1_-_31003260
|
2.431
|
NM_006762
|
LAPTM5
|
lysosomal protein transmembrane 5
|
|
chr22_-_35970230
|
2.419
|
NM_002872
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr11_-_110755112
|
2.418
|
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr16_-_30301357
|
2.404
|
|
SEPT1
|
septin 1
|
|
chr11_-_76059435
|
2.402
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr19_-_51820133
|
2.402
|
NM_000960
|
PTGIR
|
prostaglandin I2 (prostacyclin) receptor (IP)
|
|
chr1_+_196874867
|
2.393
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr7_-_44985202
|
2.388
|
NM_033054
|
MYO1G
|
myosin IG
|
|
chr18_+_70317802
|
2.358
|
NM_001168499
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr22_-_35970133
|
2.349
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr3_+_47004457
|
2.341
|
|
NBEAL2
|
neurobeachin-like 2
|
|
chrX_+_128741562
|
2.327
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr7_-_44985084
|
2.327
|
|
MYO1G
|
myosin IG
|
|
chr1_-_158815850
|
2.321
|
|
CD84
|
CD84 molecule
|
|
chr2_+_233633348
|
2.313
|
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr16_+_29581783
|
2.302
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr12_-_56422120
|
2.296
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr2_+_233633279
|
2.268
|
NM_001017915
NM_005541
|
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa
|
|
chr11_+_1830763
|
2.247
|
NM_002339
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr22_-_16080265
|
2.247
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr16_-_49272617
|
2.238
|
|
SNX20
|
sorting nexin 20
|
|
chr22_+_36008511
|
2.234
|
|
CYTH4
|
cytohesin 4
|
|
chr3_+_38055699
|
2.228
|
NM_007335
NM_007337
|
DLEC1
|
deleted in lung and esophageal cancer 1
|
|
chr12_-_56159895
|
2.222
|
NM_032496
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr12_+_12829807
|
2.216
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr17_+_44642587
|
2.203
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr1_+_157067729
|
2.202
|
NM_002432
|
MNDA
|
myeloid cell nuclear differentiation antigen
|
|
chr1_-_158815895
|
2.201
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr13_-_32658215
|
2.186
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr7_-_92615593
|
2.181
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chrX_+_128741578
|
2.177
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr2_-_242205556
|
2.171
|
NM_001164356
|
THAP4
|
THAP domain containing 4
|
|
chr15_-_78050515
|
2.169
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr8_-_82557956
|
2.167
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr19_+_41085221
|
2.145
|
NM_001007469
NM_014266
|
HCST
|
hematopoietic cell signal transducer
|
|
chr12_-_52939584
|
2.131
|
NM_001127322
|
CBX5
|
chromobox homolog 5
|
|
chr22_+_36008369
|
2.123
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chr7_-_50103335
|
2.121
|
NM_001159878
NM_007009
|
ZPBP
|
zona pellucida binding protein
|
|
chr22_+_36008465
|
2.111
|
|
CYTH4
|
cytohesin 4
|
|
chr7_+_143683365
|
2.100
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr9_+_263025
|
2.097
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr2_-_45648608
|
2.096
|
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr21_-_45165204
|
2.088
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr11_-_110755366
|
2.082
|
NM_006235
|
POU2AF1
|
POU class 2 associating factor 1
|
|
chr16_-_30301634
|
2.080
|
NM_052838
|
SEPT1
|
septin 1
|
|
chr12_-_56159631
|
2.078
|
NM_001080157
|
ARHGAP9
|
Rho GTPase activating protein 9
|
|
chr19_-_1083109
|
2.076
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr13_-_45654327
|
2.073
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr17_-_44642934
|
2.042
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr1_-_111544784
|
2.040
|
NM_024901
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr1_-_159306210
|
2.034
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr17_-_26672757
|
2.016
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr1_-_159306273
|
2.013
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr21_-_45165249
|
1.994
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr1_+_26744929
|
1.992
|
NM_001006665
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr21_+_25856311
|
1.981
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr2_+_130703384
|
1.979
|
|
|
|
|
chr19_-_11550576
|
1.977
|
NM_001111034
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr10_-_6144119
|
1.968
|
NM_000417
|
IL2RA
|
interleukin 2 receptor, alpha
|
|
chr1_+_152566899
|
1.955
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr13_-_45654296
|
1.954
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr15_-_75259714
|
1.953
|
|
PEAK1
|
NKF3 kinase family member
|
|
chr19_+_16860807
|
1.938
|
NM_003950
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr19_+_16860871
|
1.937
|
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr16_-_49272741
|
1.934
|
NM_001144972
NM_153337
NM_182854
|
SNX20
|
sorting nexin 20
|
|
chrX_-_130792297
|
1.930
|
|
LOC286467
|
hypothetical LOC286467
|
|
chr21_-_38792173
|
1.923
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_+_26744893
|
1.911
|
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1
|
|
chr6_-_154719560
|
1.907
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr1_+_170077234
|
1.897
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr14_-_93512818
|
1.897
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr16_+_30391609
|
1.896
|
|
|
|
|
chr9_-_129656413
|
1.891
|
|
ENG
|
endoglin
|
|
chr4_+_100957003
|
1.890
|
NM_014395
|
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides
|
|
chr1_+_207995999
|
1.886
|
NM_025228
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr7_+_150044530
|
1.880
|
NM_130759
|
GIMAP1
|
GTPase, IMAP family member 1
|
|
chr12_-_51887266
|
1.867
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr1_+_32489426
|
1.866
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr4_+_2783764
|
1.860
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr5_-_66528330
|
1.855
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr19_+_14412087
|
1.845
|
|
PKN1
|
protein kinase N1
|
|
chr19_+_17495109
|
1.840
|
NM_001098524
NM_173544
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr18_-_59137488
|
1.835
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr8_+_75066117
|
1.835
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr16_+_30391550
|
1.833
|
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr4_+_2783743
|
1.826
|
NM_001145855
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr2_-_179623030
|
1.821
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr17_-_59451662
|
1.809
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr9_-_129656581
|
1.803
|
|
ENG
|
endoglin
|
|
chr2_+_143603439
|
1.800
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr5_-_169657192
|
1.798
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr14_+_92188546
|
1.791
|
|
RIN3
|
Ras and Rab interactor 3
|
|
chr16_+_29582065
|
1.789
|
|
SPN
|
sialophorin
|
|
chr16_+_30391460
|
1.787
|
NM_001114380
NM_002209
|
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide)
|
|
chr19_-_15436333
|
1.783
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr17_+_4922478
|
1.762
|
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr7_-_149960612
|
1.745
|
NM_024711
|
GIMAP6
|
GTPase, IMAP family member 6
|
|
chr12_+_6517600
|
1.744
|
|
|
|
|
chr15_-_32398220
|
1.729
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr13_-_45654292
|
1.728
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr7_+_149842850
|
1.727
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr2_+_143603368
|
1.710
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr17_+_4922445
|
1.691
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr7_+_18501876
|
1.690
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr12_+_25096472
|
1.690
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr12_-_15005761
|
1.686
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr16_+_29582069
|
1.684
|
NM_003123
|
SPN
|
sialophorin
|
|
chr1_-_111544820
|
1.678
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr1_+_196874839
|
1.677
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr11_+_107304344
|
1.662
|
NM_017516
|
RAB39
|
RAB39, member RAS oncogene family
|
|
chr12_-_51887318
|
1.660
|
|
ITGB7
|
integrin, beta 7
|
|
chr2_+_130846263
|
1.658
|
|
PTPN18
|
protein tyrosine phosphatase, non-receptor type 18 (brain-derived)
|
|
chr1_+_153413675
|
1.653
|
|
TRIM46
|
tripartite motif containing 46
|
|
chr5_-_169657312
|
1.651
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr1_-_155834357
|
1.638
|
NM_031282
|
FCRL4
|
Fc receptor-like 4
|
|
chr7_-_143523582
|
1.635
|
NM_001003702
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35
|
|
chr21_-_45165302
|
1.633
|
NM_000211
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr9_-_129656778
|
1.626
|
|
ENG
|
endoglin
|
|
chr2_-_136590210
|
1.617
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr19_+_3129735
|
1.603
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr22_+_22435207
|
1.602
|
NM_182520
|
C22orf15
|
chromosome 22 open reading frame 15
|
|
chr11_+_59979857
|
1.590
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr14_-_93512647
|
1.588
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr19_-_47438583
|
1.585
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr11_+_65026382
|
1.575
|
|
|
|
|
chr22_-_35875890
|
1.574
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr2_-_175170674
|
1.566
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|