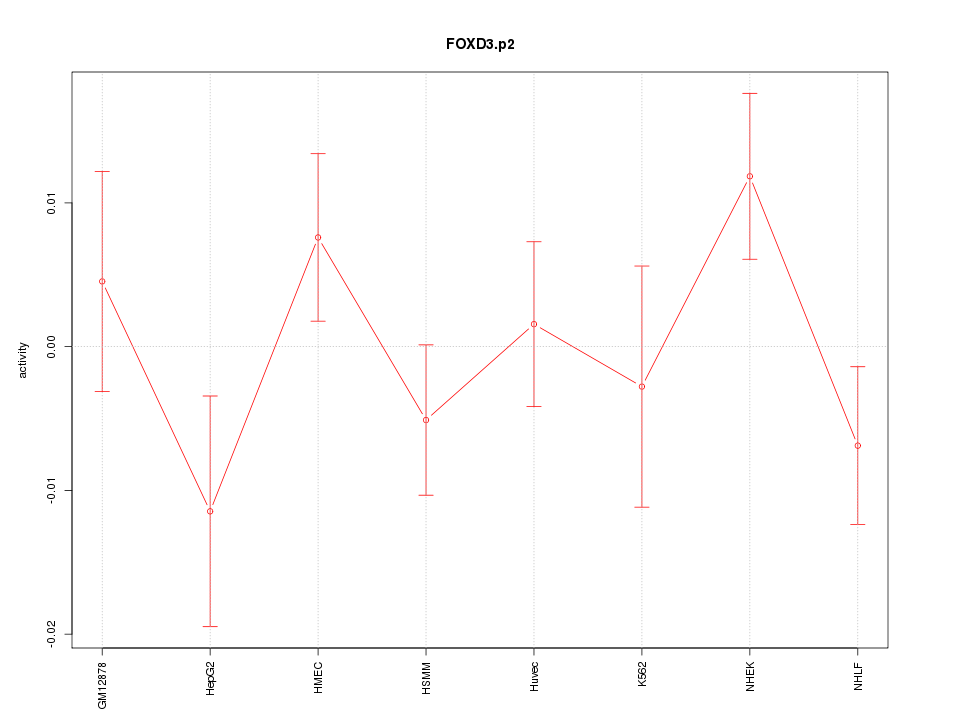
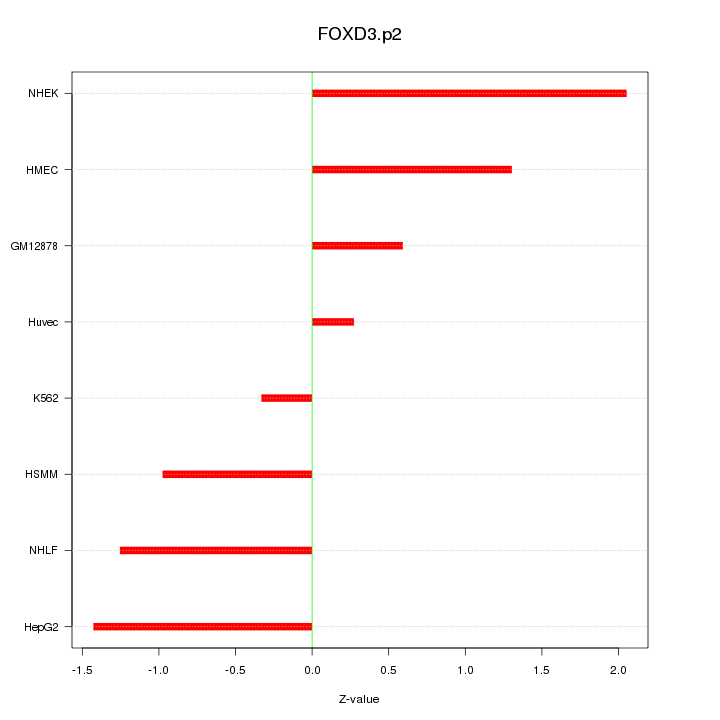
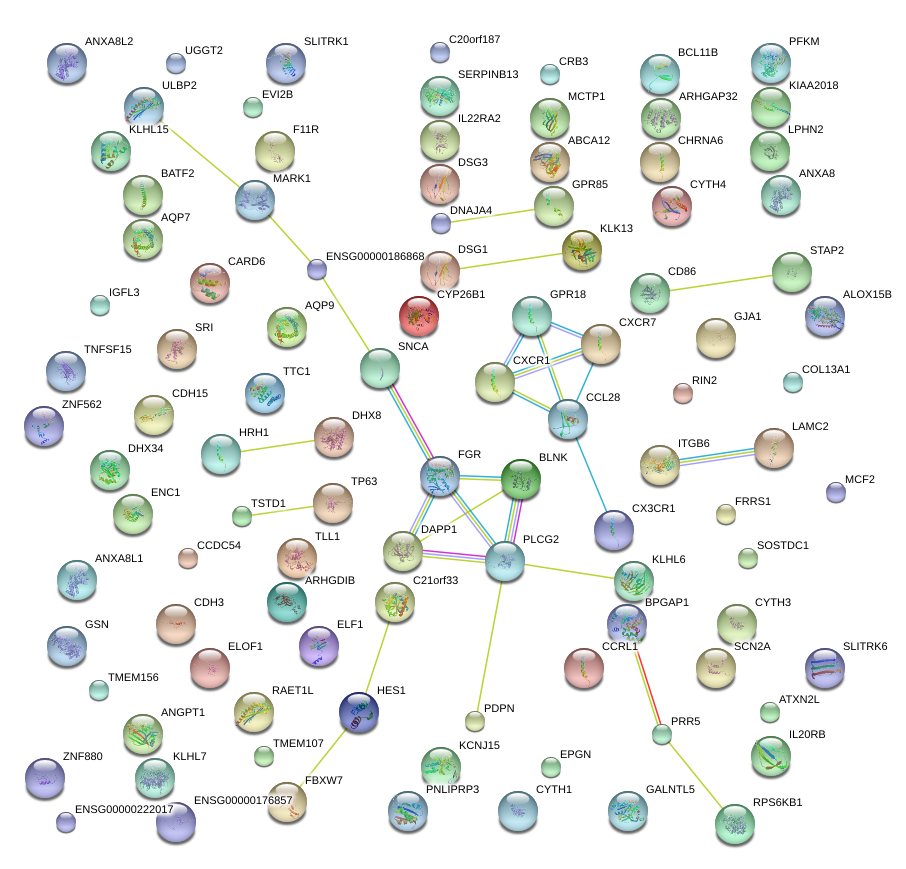
|
chr3_+_138159396
|
2.302
|
NM_144717
|
IL20RB
|
interleukin 20 receptor beta
|
|
chr18_+_59405513
|
2.105
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr4_+_75393053
|
1.921
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr20_+_19863716
|
1.877
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr13_-_40491491
|
1.672
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr18_+_27281729
|
1.640
|
NM_001944
|
DSG3
|
desmoglein 3
|
|
chr13_-_40491405
|
1.539
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr10_+_118177371
|
1.479
|
NM_001011709
|
PNLIPRP3
|
pancreatic lipase-related protein 3
|
|
chr19_+_57564981
|
1.438
|
NM_001145434
|
ZNF880
|
zinc finger protein 880
|
|
chr21_+_38550739
|
1.395
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr3_+_11242666
|
1.344
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr9_-_116608113
|
1.310
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr4_-_90978469
|
1.233
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr13_-_85271329
|
1.221
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr22_+_43527101
|
1.214
|
NM_001017526
NM_001198726
NM_181335
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr3_+_190990142
|
1.200
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr4_+_167013859
|
1.181
|
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr7_-_87687276
|
1.161
|
|
SRI
|
sorcin
|
|
chr6_+_150304813
|
1.134
|
NM_025217
|
ULBP2
|
UL16 binding protein 2
|
|
chr3_+_133798770
|
1.125
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr12_+_46799279
|
1.117
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr1_-_159275334
|
1.113
|
|
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
|
|
chr19_-_51319769
|
1.095
|
NM_207393
|
IGFL3
|
IGF-like family member 3
|
|
chr22_+_36008511
|
1.084
|
|
CYTH4
|
cytohesin 4
|
|
chrX_-_138552517
|
1.080
|
NM_001171877
NM_001171878
NM_001171879
NM_005369
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr3_-_12561962
|
1.078
|
NM_001195279
|
LOC100129480
|
hypothetical protein LOC100129480
|
|
chr14_-_98807307
|
1.058
|
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr4_-_153523113
|
1.036
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr22_+_36008465
|
0.995
|
|
CYTH4
|
cytohesin 4
|
|
chr15_+_56217686
|
0.949
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr3_+_195336625
|
0.943
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr2_-_160765008
|
0.942
|
|
ITGB6
|
integrin, beta 6
|
|
chr5_-_43432969
|
0.921
|
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr16_+_80370400
|
0.897
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr4_-_38710376
|
0.896
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr6_+_121798443
|
0.891
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr19_+_6415259
|
0.885
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chrX_-_23955223
|
0.879
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr1_-_159275357
|
0.872
|
NM_001113205
NM_001113206
NM_001113207
|
TSTD1
F11R
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1
F11 receptor
|
|
chr7_-_16471817
|
0.871
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr2_-_218739891
|
0.864
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr22_+_36008369
|
0.846
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chr4_+_100957003
|
0.844
|
NM_014395
|
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides
|
|
chr1_+_181421796
|
0.832
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr1_+_13784553
|
0.809
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr5_+_159368690
|
0.802
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr11_-_128399218
|
0.795
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr2_-_215711313
|
0.789
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr6_-_137536412
|
0.787
|
NM_052962
NM_181309
NM_181310
|
IL22RA2
|
interleukin 22 receptor, alpha 2
|
|
chr5_+_159368733
|
0.785
|
NM_003314
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr1_-_100003936
|
0.775
|
NM_001013660
|
FRRS1
|
ferric-chelate reductase 1
|
|
chr6_+_49626071
|
0.768
|
NM_001145652
|
C6orf141
|
chromosome 6 open reading frame 141
|
|
chr7_-_112513379
|
0.762
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr7_-_87687302
|
0.756
|
NM_003130
|
SRI
|
sorcin
|
|
chr7_+_151284396
|
0.746
|
NM_145292
|
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5
|
|
chr1_+_81544432
|
0.738
|
|
LPHN2
|
latrophilin 2
|
|
chr2_+_165858586
|
0.724
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr13_-_83354474
|
0.711
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr8_-_42742729
|
0.703
|
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr15_+_76345577
|
0.703
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr10_-_98021144
|
0.697
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr17_+_7883076
|
0.691
|
NM_001039130
NM_001039131
NM_001141
|
ALOX15B
|
arachidonate 15-lipoxygenase, type B
|
|
chr8_-_108579270
|
0.683
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr13_-_83354425
|
0.682
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr3_-_184756101
|
0.677
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr10_+_47875229
|
0.676
|
NM_001040084
NM_001098845
|
ANXA8L2
ANXA8
ANXA8L1
|
annexin A8-like 2
annexin A8
annexin A8-like 1
|
|
chr2_+_237143118
|
0.676
|
NM_020311
|
CXCR7
|
chemokine (C-X-C motif) receptor 7
|
|
chr3_+_123256902
|
0.672
|
NM_175862
|
CD86
|
CD86 molecule
|
|
chr5_-_94442941
|
0.671
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr13_-_98708549
|
0.666
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr1_-_27825178
|
0.652
|
NM_001042747
|
FGR
|
Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog
|
|
chr10_+_47216925
|
0.652
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr2_-_72228427
|
0.648
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr19_-_4289750
|
0.645
|
NM_001013841
NM_017720
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr1_+_13782838
|
0.643
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr16_+_67235651
|
0.643
|
NM_001793
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr10_-_46593923
|
0.636
|
NM_001040084
NM_001098845
|
ANXA8L2
LOC653110
ANXA8
ANXA8L1
|
annexin A8-like 2
hypothetical LOC653110
annexin A8
annexin A8-like 1
|
|
chr3_+_108578877
|
0.634
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr22_+_44828338
|
0.627
|
|
|
|
|
chr5_+_40877053
|
0.627
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr1_+_218768170
|
0.625
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr17_-_26665213
|
0.610
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr12_-_15005761
|
0.604
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr10_+_71231693
|
0.603
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr18_+_27152049
|
0.603
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr1_+_190811479
|
0.596
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr20_+_51023159
|
0.595
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr1_-_77457664
|
0.595
|
NM_005482
|
PIGK
|
phosphatidylinositol glycan anchor biosynthesis, class K
|
|
chr1_-_158948176
|
0.593
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr17_-_26665194
|
0.592
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr5_+_159368721
|
0.581
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr1_+_179719338
|
0.578
|
NM_000721
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr12_+_55201854
|
0.575
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr5_-_59100194
|
0.573
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_27066749
|
0.571
|
NM_004264
|
MED21
|
mediator complex subunit 21
|
|
chr17_-_26665144
|
0.569
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr19_-_4782711
|
0.568
|
NM_182919
|
TICAM1
|
toll-like receptor adaptor molecule 1
|
|
chr17_-_26665220
|
0.564
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr14_-_60817946
|
0.562
|
|
|
|
|
chr12_+_27066768
|
0.558
|
|
MED21
|
mediator complex subunit 21
|
|
chr21_+_38550533
|
0.557
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr3_+_195336631
|
0.540
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr12_-_83954185
|
0.539
|
NM_001100917
|
TSPAN19
|
tetraspanin 19
|
|
chr2_-_160764819
|
0.531
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr7_-_92615593
|
0.531
|
NM_152703
|
SAMD9L
|
sterile alpha motif domain containing 9-like
|
|
chr3_-_191321367
|
0.531
|
|
LEPREL1
|
leprecan-like 1
|
|
chr6_+_31190505
|
0.527
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr15_-_58477407
|
0.526
|
NM_001002857
NM_001002858
NM_001136015
NM_004039
|
ANXA2
|
annexin A2
|
|
chr17_-_8364942
|
0.525
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr19_+_42689881
|
0.524
|
|
ZNF793
|
zinc finger protein 793
|
|
chr19_-_56946911
|
0.524
|
NM_001193306
NM_002029
|
FPR1
|
formyl peptide receptor 1
|
|
chr18_-_33399601
|
0.523
|
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr21_-_41140457
|
0.520
|
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr12_+_29193399
|
0.517
|
|
FAR2
|
fatty acyl CoA reductase 2
|
|
chr14_-_60818263
|
0.512
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr10_+_71231649
|
0.511
|
NM_001130103
NM_080798
NM_080800
NM_080801
NM_080802
NM_080805
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr2_+_201182187
|
0.510
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr2_-_158008763
|
0.510
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr19_-_11550765
|
0.504
|
NM_001111035
NM_001111036
|
ACP5
|
acid phosphatase 5, tartrate resistant
|
|
chr19_+_3087026
|
0.486
|
NM_002068
|
GNA15
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class)
|
|
chr3_+_123385219
|
0.485
|
NM_000388
|
CASR
|
calcium-sensing receptor
|
|
chr2_-_213723298
|
0.484
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr2_-_108972048
|
0.481
|
|
EDAR
|
ectodysplasin A receptor
|
|
chr1_+_218768146
|
0.474
|
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr19_-_10558906
|
0.473
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr19_-_19793512
|
0.472
|
NM_001099269
NM_001145404
|
ZNF506
|
zinc finger protein 506
|
|
chr7_-_20223467
|
0.466
|
NM_182762
|
MACC1
|
metastasis associated in colon cancer 1
|
|
chr10_-_116241572
|
0.461
|
|
|
|
|
chr21_-_38792173
|
0.458
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr22_-_23500682
|
0.454
|
NM_001008496
|
PIWIL3
|
piwi-like 3 (Drosophila)
|
|
chr14_-_80475636
|
0.454
|
NM_152446
|
C14orf145
|
chromosome 14 open reading frame 145
|
|
chr3_+_123779122
|
0.452
|
NM_001113523
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr11_-_26550243
|
0.452
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr1_-_21373816
|
0.450
|
NM_001198803
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3
|
|
chr17_-_53847937
|
0.450
|
|
RNF43
|
ring finger protein 43
|
|
chr2_+_102320148
|
0.446
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr3_+_123526700
|
0.445
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr19_+_59820413
|
0.444
|
NM_006669
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr19_+_45973139
|
0.439
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr3_-_112796855
|
0.435
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr1_+_34993234
|
0.434
|
NM_005268
|
GJB5
|
gap junction protein, beta 5, 31.1kDa
|
|
chr3_+_29297806
|
0.433
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr5_+_96237923
|
0.433
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr6_+_26348539
|
0.432
|
NM_003540
|
HIST1H4F
|
histone cluster 1, H4f
|
|
chr2_-_85490997
|
0.432
|
NM_001747
|
CAPG
|
capping protein (actin filament), gelsolin-like
|
|
chr9_-_25668230
|
0.431
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr1_+_107485071
|
0.430
|
|
NTNG1
|
netrin G1
|
|
chr14_+_101345878
|
0.428
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr12_+_21175394
|
0.428
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr19_+_48772791
|
0.424
|
NM_001193621
|
LOC390940
|
hypothetical protein LOC390940
|
|
chr21_+_29439768
|
0.423
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr6_-_32892705
|
0.423
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr2_-_108972100
|
0.422
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr1_+_84540921
|
0.421
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr1_+_104093962
|
0.415
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr1_+_158975662
|
0.410
|
NM_021181
|
SLAMF7
|
SLAM family member 7
|
|
chrX_-_101297417
|
0.406
|
NM_001159560
NM_001012978
|
BEX5
|
brain expressed, X-linked 5
|
|
chr9_+_676640
|
0.404
|
|
|
|
|
chr12_+_77782579
|
0.404
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr1_-_221920023
|
0.401
|
NM_001143962
|
CAPN8
|
calpain 8
|
|
chr9_-_87904844
|
0.401
|
NM_177937
|
GOLM1
|
golgi membrane protein 1
|
|
chr4_-_153820535
|
0.399
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr3_-_188339882
|
0.397
|
NM_052969
|
RPL39L
|
ribosomal protein L39-like
|
|
chr4_-_38534802
|
0.397
|
NM_006068
|
TLR6
|
toll-like receptor 6
|
|
chr3_+_137224266
|
0.395
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr5_-_24680841
|
0.393
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr7_-_102070473
|
0.390
|
NM_001114403
|
UPK3BL
POLR2J2
|
uroplakin 3B-like
polymerase (RNA) II (DNA directed) polypeptide J2
|
|
chr19_+_42689680
|
0.386
|
NM_001013659
|
ZNF793
|
zinc finger protein 793
|
|
chr11_+_112763722
|
0.385
|
NM_178510
|
ANKK1
|
ankyrin repeat and kinase domain containing 1
|
|
chr12_+_52708460
|
0.380
|
NM_004503
|
HOXC6
|
homeobox C6
|
|
chr1_-_177378801
|
0.377
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr12_+_52696908
|
0.377
|
NM_014620
NM_153693
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr1_-_158759606
|
0.374
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chr10_-_116276560
|
0.373
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr7_+_28692122
|
0.370
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_-_83116414
|
0.369
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr18_+_59705918
|
0.367
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr1_-_221919982
|
0.366
|
|
CAPN8
|
calpain 8
|
|
chr9_+_27514311
|
0.366
|
NM_020124
|
IFNK
|
interferon, kappa
|
|
chr2_-_121759237
|
0.365
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr5_+_176717449
|
0.359
|
NM_006480
|
RGS14
|
regulator of G-protein signaling 14
|
|
chr4_+_75449720
|
0.359
|
NM_001432
|
EREG
|
epiregulin
|
|
chr13_-_74799898
|
0.359
|
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr9_+_74956601
|
0.355
|
|
ANXA1
|
annexin A1
|
|
chr11_+_67107852
|
0.353
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr3_-_15814678
|
0.352
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr3_+_179736917
|
0.352
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr16_-_28542366
|
0.348
|
NM_177536
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr3_-_170346760
|
0.348
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chrX_+_11221453
|
0.346
|
NM_001142
NM_182680
NM_182681
|
AMELX
|
amelogenin, X-linked
|
|
chr5_+_36644187
|
0.346
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr15_-_43458247
|
0.345
|
NM_001482
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr9_+_126460511
|
0.344
|
|
|
|
|
chr9_+_5500544
|
0.341
|
NM_025239
|
PDCD1LG2
|
programmed cell death 1 ligand 2
|
|
chr19_-_55539769
|
0.341
|
|
NAPSB
|
napsin B aspartic peptidase pseudogene
|
|
chr12_-_28016182
|
0.338
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr11_-_107969554
|
0.335
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr5_-_58607673
|
0.335
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_36719030
|
0.335
|
NM_001146182
|
KRTAP16-1
|
keratin associated protein 16-1
|
|
chr3_+_100840129
|
0.333
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|