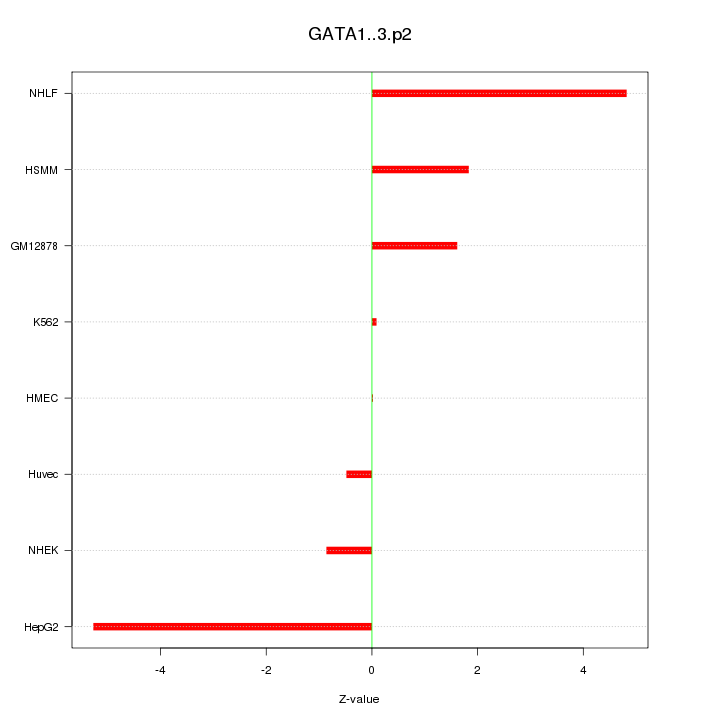
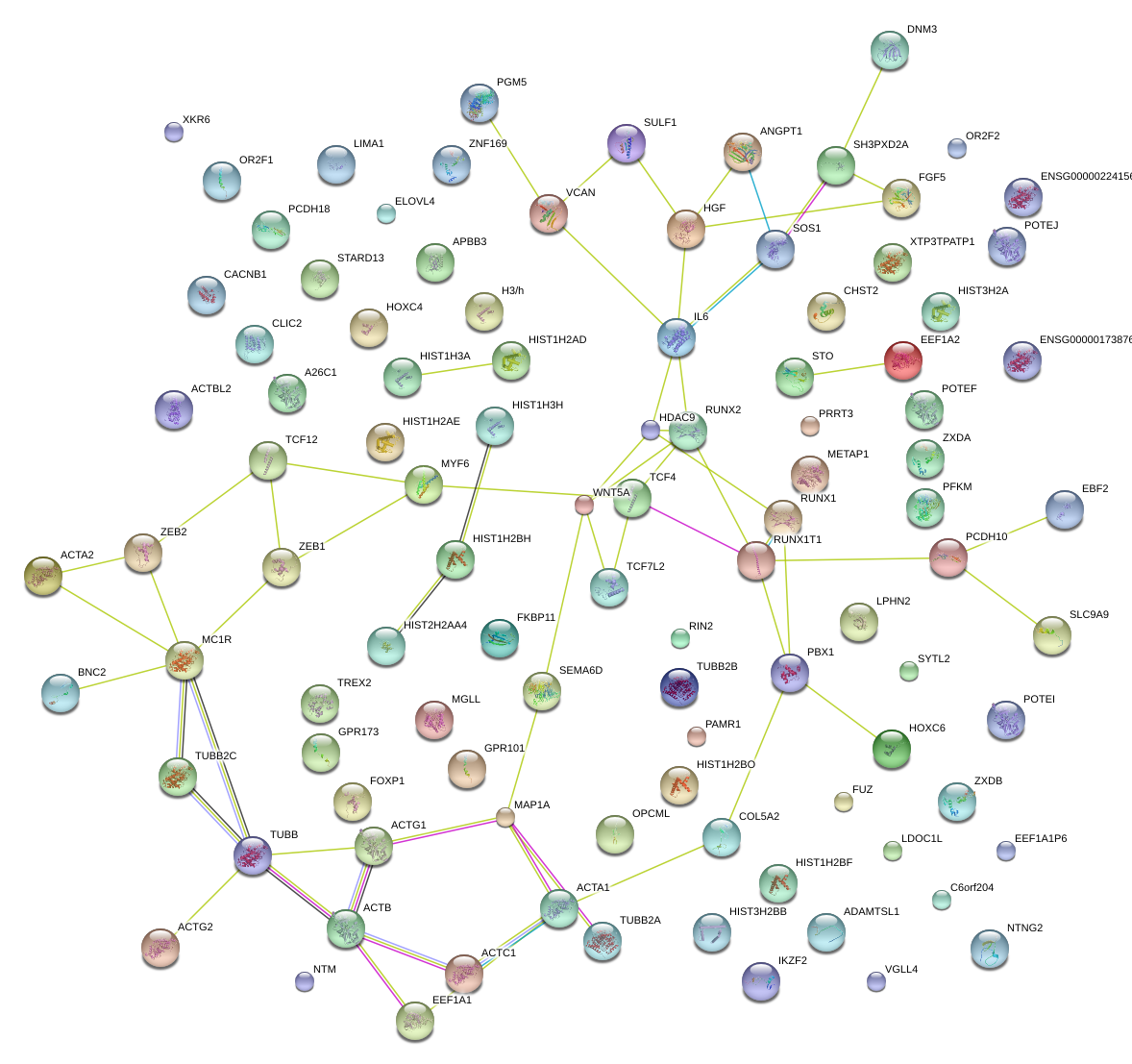
|
chr7_-_5534252
|
2.746
|
|
ACTB
|
actin, beta
|
|
chr12_-_48902692
|
2.498
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chrX_+_53095230
|
2.474
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr2_-_189752575
|
2.415
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr7_-_81237162
|
2.318
|
NM_000601
NM_001010931
NM_001010932
NM_001010933
NM_001010934
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor)
|
|
chr6_-_74284522
|
2.272
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr8_-_108579270
|
2.209
|
NM_001146
|
ANGPT1
|
angiopoietin 1
|
|
chr3_-_129023850
|
1.969
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr7_-_5534737
|
1.925
|
|
ACTB
|
actin, beta
|
|
chr8_-_11096257
|
1.890
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr8_-_93176819
|
1.866
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_-_35503720
|
1.866
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr6_+_30799130
|
1.823
|
|
TUBB
|
tubulin, beta
|
|
chr4_-_138673078
|
1.760
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr1_+_81544432
|
1.745
|
|
LPHN2
|
latrophilin 2
|
|
chr11_-_85107569
|
1.742
|
NM_206929
|
SYTL2
|
synaptotagmin-like 2
|
|
chr19_-_55008172
|
1.703
|
NM_001171937
NM_025129
|
FUZ
|
fuzzy homolog (Drosophila)
|
|
chr20_+_19903778
|
1.693
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr3_+_144321286
|
1.634
|
NM_004267
|
CHST2
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2
|
|
chr9_-_16717887
|
1.615
|
|
BNC2
|
basonuclin 2
|
|
chr9_+_134026836
|
1.577
|
|
NTNG2
|
netrin G2
|
|
chr10_-_105427898
|
1.570
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr4_+_134292630
|
1.565
|
|
PCDH10
|
protocadherin 10
|
|
chr8_-_93176618
|
1.553
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_140871107
|
1.537
|
|
|
|
|
chr15_+_41597097
|
1.476
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr4_-_138673014
|
1.447
|
|
PCDH18
|
protocadherin 18
|
|
chr6_-_119137923
|
1.404
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr4_+_134293171
|
1.389
|
|
PCDH10
|
protocadherin 10
|
|
chr2_-_213723298
|
1.380
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr7_+_18501876
|
1.358
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr3_-_145049853
|
1.351
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr1_+_162795538
|
1.348
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr12_+_52733927
|
1.346
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr1_-_226712140
|
1.341
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr6_-_26379567
|
1.329
|
NM_003534
|
HIST1H3G
|
histone cluster 1, H3g
|
|
chr8_+_70541631
|
1.310
|
|
SULF1
|
sulfatase 1
|
|
chr8_-_93144366
|
1.302
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93144039
|
1.285
|
|
|
|
|
chrX_-_154216929
|
1.284
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr8_-_93099040
|
1.276
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_140870881
|
1.260
|
|
|
|
|
chr1_+_226712430
|
1.258
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr22_-_43272100
|
1.230
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr18_-_51219880
|
1.206
|
|
TCF4
|
transcription factor 4
|
|
chr15_+_45797977
|
1.175
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chrX_-_135941498
|
1.158
|
NM_054021
|
GPR101
|
G protein-coupled receptor 101
|
|
chr5_+_176494690
|
1.148
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr11_+_131285747
|
1.147
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr12_+_46799279
|
1.130
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr8_-_25958291
|
1.122
|
NM_022659
|
EBF2
|
early B-cell factor 2
|
|
chr9_+_70161634
|
1.119
|
NM_021965
|
PGM5
|
phosphoglucomutase 5
|
|
chr20_-_56859352
|
1.118
|
|
GNAS-AS1
|
GNAS antisense RNA 1 (non-protein coding)
|
|
chr1_+_170077234
|
1.110
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr2_-_144994349
|
1.086
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr3_-_9969057
|
1.074
|
NM_207351
|
PRRT3
|
proline-rich transmembrane protein 3
|
|
chr9_+_18464138
|
1.069
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr4_+_81406765
|
1.058
|
NM_004464
NM_033143
|
FGF5
|
fibroblast growth factor 5
|
|
chr5_-_139924229
|
1.043
|
NM_006051
NM_133172
NM_133173
NM_133174
|
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3
|
|
chr7_+_143263191
|
1.040
|
NM_001004685
|
OR2F2
|
olfactory receptor, family 2, subfamily F, member 2
|
|
chr2_-_144994038
|
1.036
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr17_-_45629401
|
1.025
|
|
|
|
|
chr5_+_82803248
|
1.011
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr6_-_80713589
|
0.998
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr15_+_55298908
|
0.995
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chr17_-_45626364
|
0.991
|
|
|
|
|
chr12_+_79625538
|
0.989
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chrX_-_57953578
|
0.982
|
NM_007156
|
ZXDA
|
zinc finger, X-linked, duplicated A
|
|
chr13_-_32678142
|
0.979
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr21_-_35153664
|
0.969
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_11598829
|
0.964
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr17_-_77092569
|
0.962
|
|
ACTG1
|
actin, gamma 1
|
|
chr16_+_88305743
|
0.957
|
|
LOC100128881
|
hypothetical LOC100128881
|
|
chr3_-_71262426
|
0.956
|
|
FOXP1
|
forkhead box P1
|
|
chr11_-_85108030
|
0.951
|
NM_001162952
NM_206930
|
SYTL2
|
synaptotagmin-like 2
|
|
chr3_-_55489973
|
0.951
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr9_+_134027154
|
0.946
|
NM_032536
|
NTNG2
|
netrin G2
|
|
chr5_-_158459276
|
0.934
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr3_+_179736917
|
0.928
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr17_-_45633873
|
0.922
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr10_-_79067210
|
0.920
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr18_-_50004458
|
0.915
|
|
MBD2
|
methyl-CpG binding domain protein 2
|
|
chr5_-_88214720
|
0.915
|
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_-_18585721
|
0.910
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr9_+_76302071
|
0.904
|
NM_006914
|
RORB
|
RAR-related orphan receptor B
|
|
chr19_-_45888259
|
0.895
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr3_+_142587973
|
0.893
|
|
|
|
|
chr8_+_70541412
|
0.893
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr6_+_33151703
|
0.881
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr6_-_130224108
|
0.879
|
NM_001010876
|
C6orf191
|
chromosome 6 open reading frame 191
|
|
chr1_+_162795328
|
0.876
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr2_+_157822355
|
0.869
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr10_+_75343352
|
0.862
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr1_+_200775864
|
0.859
|
NM_001197131
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr3_-_113841448
|
0.848
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr8_-_93177057
|
0.847
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_144991386
|
0.847
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr1_-_26517342
|
0.843
|
NM_145345
|
UBXN11
|
UBX domain protein 11
|
|
chr8_-_25958556
|
0.834
|
|
EBF2
|
early B-cell factor 2
|
|
chr9_+_123501186
|
0.832
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr4_+_160408447
|
0.831
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr12_+_103375116
|
0.828
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chrX_+_51503219
|
0.823
|
NM_018094
|
GSPT2
|
G1 to S phase transition 2
|
|
chr3_-_71262405
|
0.822
|
|
FOXP1
|
forkhead box P1
|
|
chr8_+_70567634
|
0.820
|
|
SULF1
|
sulfatase 1
|
|
chr17_-_44641741
|
0.819
|
NM_031498
NM_001198755
NM_001198756
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr12_-_15005761
|
0.815
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr1_+_66772393
|
0.814
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr9_+_18464090
|
0.814
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr9_-_35608300
|
0.812
|
NM_001782
|
CD72
|
CD72 molecule
|
|
chr1_-_72520735
|
0.809
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr17_+_39990124
|
0.804
|
NM_001466
|
FZD2
|
frizzled homolog 2 (Drosophila)
|
|
chr14_+_95792299
|
0.803
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr11_-_62233552
|
0.798
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr5_+_67558217
|
0.797
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr8_+_70567581
|
0.797
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr5_+_139924224
|
0.792
|
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr15_-_81667169
|
0.789
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr2_-_144991556
|
0.789
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr13_-_101852028
|
0.787
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr11_+_5610060
|
0.782
|
NM_130390
|
TRIM34
|
tripartite motif containing 34
|
|
chr11_-_47427263
|
0.780
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr2_-_39040981
|
0.780
|
|
LOC375196
|
hypothetical LOC375196
|
|
chr1_-_153509473
|
0.779
|
NM_003993
|
CLK2
|
CDC-like kinase 2
|
|
chr11_-_95715976
|
0.769
|
NM_032427
|
MAML2
|
mastermind-like 2 (Drosophila)
|
|
chr6_-_78229838
|
0.764
|
NM_000863
|
HTR1B
|
5-hydroxytryptamine (serotonin) receptor 1B
|
|
chr10_-_79067552
|
0.762
|
NM_001014797
NM_001161352
NM_001161353
NM_002247
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr10_-_79067476
|
0.757
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr3_-_55496607
|
0.754
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr7_+_90176647
|
0.753
|
NM_012395
|
CDK14
|
cyclin-dependent kinase 14
|
|
chr16_-_87570716
|
0.753
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr18_-_65766068
|
0.750
|
|
CD226
|
CD226 molecule
|
|
chr7_+_128795178
|
0.748
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr13_-_105985313
|
0.748
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr6_-_101018271
|
0.744
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr15_+_39032927
|
0.743
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr1_+_234372454
|
0.743
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr5_-_124108672
|
0.741
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr12_+_79634820
|
0.739
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr6_+_26128656
|
0.738
|
NM_003529
|
HIST1H3A
|
histone cluster 1, H3a
|
|
chr15_-_78050515
|
0.731
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr1_+_154095883
|
0.729
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr17_-_44022595
|
0.727
|
|
HOXB3
|
homeobox B3
|
|
chr10_+_18669619
|
0.725
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr10_+_70517863
|
0.712
|
|
SRGN
|
serglycin
|
|
chrX_+_78087484
|
0.712
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr11_-_133332089
|
0.711
|
NM_014987
|
IGSF9B
|
immunoglobulin superfamily, member 9B
|
|
chr2_-_38156769
|
0.709
|
|
CYP1B1
|
cytochrome P450, family 1, subfamily B, polypeptide 1
|
|
chr5_+_36644187
|
0.703
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr12_+_77782719
|
0.702
|
|
SYT1
|
synaptotagmin I
|
|
chr9_+_112470871
|
0.701
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr3_+_49002774
|
0.699
|
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr10_+_70517823
|
0.696
|
NM_002727
|
SRGN
|
serglycin
|
|
chr3_+_49002285
|
0.693
|
NM_177938
NM_177939
|
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum)
|
|
chr7_+_128795280
|
0.690
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr10_-_79067400
|
0.689
|
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr6_-_76259976
|
0.677
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr1_+_207823667
|
0.670
|
NM_020439
|
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG
|
|
chr17_+_7199178
|
0.670
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chr12_+_79362256
|
0.662
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr13_+_100902942
|
0.662
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr1_-_85816520
|
0.657
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_+_189547358
|
0.657
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_+_50318801
|
0.650
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr10_-_105202139
|
0.645
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr13_-_107317460
|
0.644
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr17_-_44026042
|
0.639
|
NM_002147
|
HOXB5
|
homeobox B5
|
|
chr1_-_42157077
|
0.635
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr5_+_140733664
|
0.635
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr4_+_75449720
|
0.634
|
NM_001432
|
EREG
|
epiregulin
|
|
chr14_+_64077171
|
0.632
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr22_-_35970133
|
0.631
|
|
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2)
|
|
chr12_-_52281050
|
0.631
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr17_-_43977273
|
0.629
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr12_-_51912101
|
0.629
|
NM_000966
|
RARG
|
retinoic acid receptor, gamma
|
|
chr6_+_6533932
|
0.627
|
NM_004271
|
LY86
|
lymphocyte antigen 86
|
|
chr11_+_63730720
|
0.626
|
NM_031471
NM_178443
|
FERMT3
|
fermitin family member 3
|
|
chr2_-_167058829
|
0.626
|
|
SCN7A
|
sodium channel, voltage-gated, type VII, alpha
|
|
chr10_-_105202012
|
0.624
|
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr22_-_43271843
|
0.623
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr3_-_113841200
|
0.623
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr14_+_101346032
|
0.619
|
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr12_+_77782579
|
0.615
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr1_-_42156913
|
0.614
|
|
|
|
|
chr15_-_54322767
|
0.614
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr3_-_55496370
|
0.613
|
NM_003392
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr3_+_99733432
|
0.612
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr2_+_189547323
|
0.611
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_+_120756325
|
0.611
|
NM_057168
|
WNT16
|
wingless-type MMTV integration site family, member 16
|
|
chr2_-_215711313
|
0.610
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr12_+_56206692
|
0.608
|
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr2_-_167058901
|
0.607
|
|
SCN7A
|
sodium channel, voltage-gated, type VII, alpha
|
|
chr2_+_189547377
|
0.597
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr10_-_75005421
|
0.593
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr2_-_29150630
|
0.592
|
NM_001029883
|
C2orf71
|
chromosome 2 open reading frame 71
|
|
chr11_+_58146721
|
0.590
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr10_-_100985548
|
0.588
|
NM_001166244
NM_001166245
NM_001166246
NM_021828
|
HPSE2
|
heparanase 2
|
|
chr8_-_116750427
|
0.588
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr17_-_77092831
|
0.587
|
|
ACTG1
|
actin, gamma 1
|
|
chr3_+_190831909
|
0.587
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|