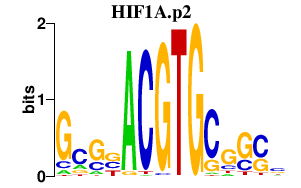
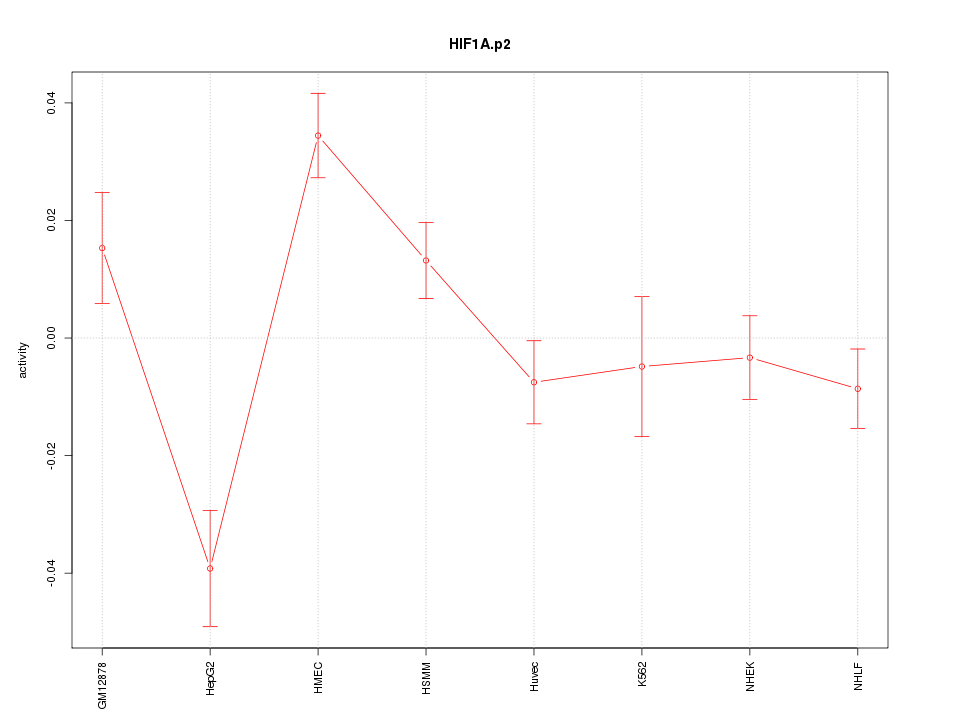
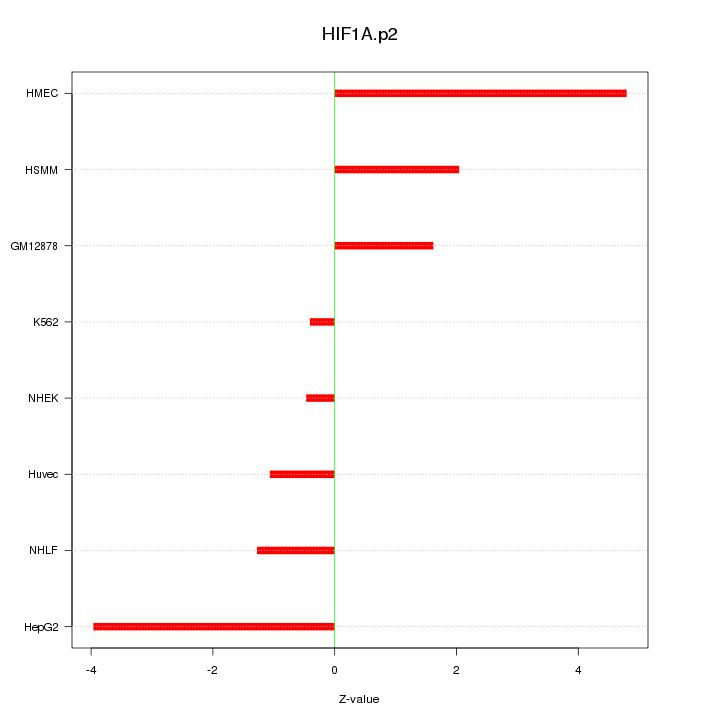
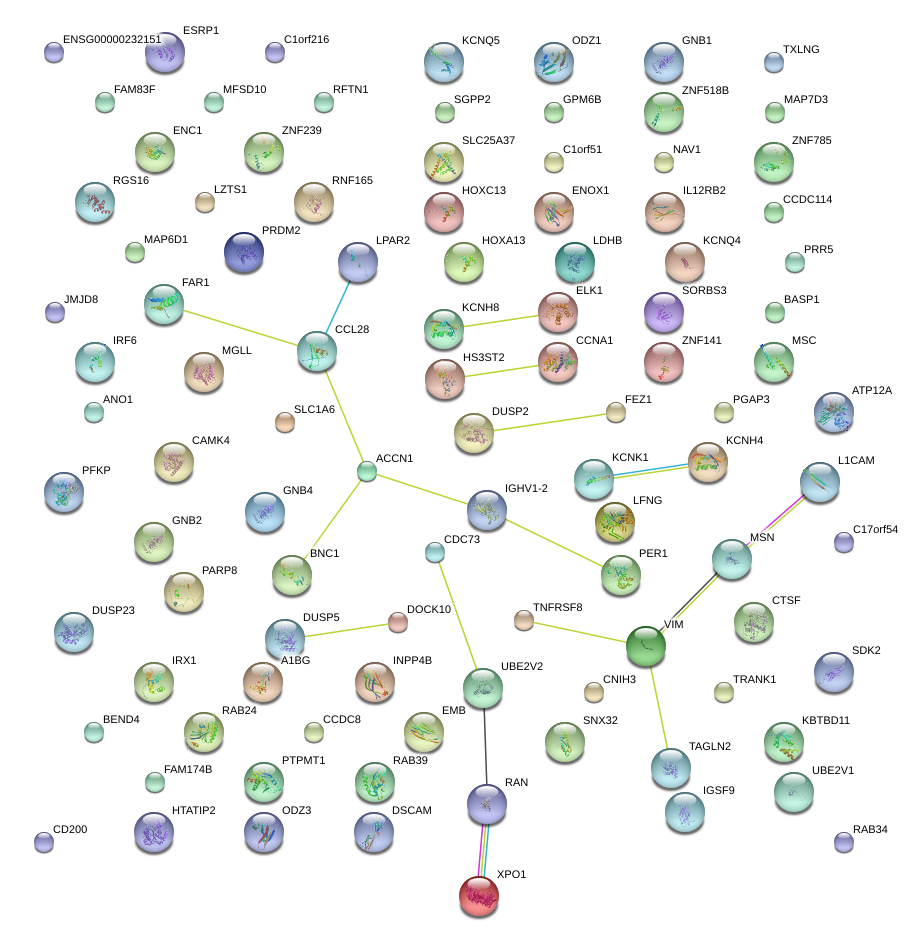
|
chr22_+_38720881
|
3.360
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chrX_-_13866366
|
2.528
|
|
|
|
|
chrX_-_13866565
|
2.421
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chrX_-_13866451
|
2.357
|
|
GPM6B
|
glycoprotein M6B
|
|
chr3_-_185025989
|
2.251
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr13_-_43259032
|
1.982
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr19_-_51608665
|
1.838
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr2_-_96174502
|
1.759
|
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr11_+_69602055
|
1.654
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr1_+_231816372
|
1.512
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr4_-_41848874
|
1.481
|
|
BEND4
|
BEN domain containing 4
|
|
chr10_-_88720908
|
1.478
|
|
LOC100133190
|
hypothetical LOC100133190
|
|
chr4_+_321607
|
1.419
|
|
ZNF141
|
zinc finger protein 141
|
|
chr6_+_73388713
|
1.418
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr19_-_63550712
|
1.403
|
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr4_+_321587
|
1.379
|
NM_003441
|
ZNF141
|
zinc finger protein 141
|
|
chr2_-_96174865
|
1.373
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr1_+_13899272
|
1.372
|
NM_001135610
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr5_+_17270781
|
1.368
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr5_+_3649167
|
1.351
|
NM_024337
|
IRX1
|
iroquois homeobox 1
|
|
chr19_-_19599973
|
1.348
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr2_-_225615556
|
1.344
|
NM_014689
|
DOCK10
|
dedicator of cytokinesis 10
|
|
chr12_+_52618815
|
1.321
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr21_+_25856311
|
1.288
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chr4_-_10068055
|
1.141
|
NM_053042
|
ZNF518B
|
zinc finger protein 518B
|
|
chr1_-_180840016
|
1.129
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr5_+_17270474
|
1.103
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr11_+_65357972
|
1.086
|
NM_152760
|
SNX32
|
sorting nexin 32
|
|
chr1_+_231816540
|
1.085
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr3_-_180651874
|
1.068
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr3_-_129024665
|
1.067
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr2_+_222997456
|
1.054
|
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr10_+_112247614
|
1.035
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr2_+_222997529
|
1.034
|
NM_152386
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr5_-_49772940
|
0.997
|
|
EMB
|
embigin
|
|
chr8_+_95722448
|
0.982
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr5_-_49772973
|
0.978
|
NM_198449
|
EMB
|
embigin
|
|
chr13_+_24152548
|
0.968
|
NM_001185085
NM_001676
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide
|
|
chr3_-_129023850
|
0.964
|
NM_007283
|
MGLL
|
monoglyceride lipase
|
|
chr12_+_52806103
|
0.960
|
|
LOC400043
|
hypothetical LOC400043
|
|
chr4_+_183302105
|
0.929
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr12_-_21701985
|
0.925
|
NM_001174097
|
LDHB
|
lactate dehydrogenase B
|
|
chr11_+_13646806
|
0.923
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr3_-_180652024
|
0.919
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr12_-_21702021
|
0.918
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21702036
|
0.909
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr12_-_21702000
|
0.901
|
|
LDHB
|
lactate dehydrogenase B
|
|
chr11_+_13646727
|
0.900
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr3_+_113534710
|
0.890
|
|
CD200
|
CD200 molecule
|
|
chr10_+_17312654
|
0.868
|
|
VIM
|
vimentin
|
|
chr11_-_66092534
|
0.868
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr1_+_222870573
|
0.867
|
|
CNIH3
|
cornichon homolog 3 (Drosophila)
|
|
chr11_+_20341835
|
0.852
|
NM_001098520
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr22_+_16020241
|
0.851
|
|
CECR4
|
cat eye syndrome chromosome region, candidate 4 (non-protein coding)
|
|
chr11_+_20341806
|
0.850
|
NM_006410
NM_001098521
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr17_-_7996409
|
0.849
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr1_+_222870798
|
0.848
|
NM_152495
|
CNIH3
|
cornichon homolog 3 (Drosophila)
|
|
chr11_-_124871258
|
0.847
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr1_-_35957030
|
0.844
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr4_-_143987053
|
0.819
|
NM_001101669
NM_003866
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa
|
|
chr1_-_208046011
|
0.818
|
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr12_-_21702046
|
0.811
|
NM_002300
|
LDHB
|
lactate dehydrogenase B
|
|
chrX_-_135161185
|
0.808
|
NM_001173517
NM_024597
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr10_+_3099708
|
0.807
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr16_-_674357
|
0.801
|
NM_001005920
|
JMJD8
|
jumonji domain containing 8
|
|
chr18_+_42167904
|
0.801
|
|
RNF165
|
ring finger protein 165
|
|
chr1_+_148521852
|
0.800
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr8_+_1937711
|
0.795
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr1_-_35956979
|
0.789
|
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr13_+_35903966
|
0.789
|
NM_001111046
NM_001111047
|
CCNA1
|
cyclin A1
|
|
chr5_+_110587546
|
0.777
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr16_-_674318
|
0.775
|
|
JMJD8
|
jumonji domain containing 8
|
|
chr8_+_22479115
|
0.774
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr11_+_20341900
|
0.767
|
NM_001098522
|
HTATIP2
|
HIV-1 Tat interactive protein 2, 30kDa
|
|
chr19_-_14951349
|
0.761
|
|
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6
|
|
chr8_-_72918697
|
0.759
|
|
MSC
|
musculin
|
|
chrX_+_64804235
|
0.759
|
NM_002444
|
MSN
|
moesin
|
|
chr1_+_158017380
|
0.753
|
NM_017823
|
DUSP23
|
dual specificity phosphatase 23
|
|
chr3_-_36961539
|
0.749
|
NM_014831
|
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr17_-_69151821
|
0.739
|
NM_001144952
|
SDK2
|
sidekick homolog 2 (chicken)
|
|
chr15_-_90999606
|
0.727
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr21_-_41140457
|
0.725
|
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr11_+_107304344
|
0.720
|
NM_017516
|
RAB39
|
RAB39, member RAS oncogene family
|
|
chr1_+_191357906
|
0.711
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr16_+_22733307
|
0.700
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chrX_-_47394808
|
0.699
|
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chrX_+_16714452
|
0.698
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chrX_-_47394937
|
0.697
|
NM_001114123
NM_005229
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chrX_-_152804731
|
0.697
|
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr15_-_81744469
|
0.694
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr19_-_53516962
|
0.686
|
|
CCDC114
|
coiled-coil domain containing 114
|
|
chr1_+_12046020
|
0.683
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr1_-_158182006
|
0.679
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr22_+_43476710
|
0.675
|
NM_181333
|
PRR5
|
proline rich 5 (renal)
|
|
chr7_+_2525922
|
0.674
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr16_-_30504309
|
0.672
|
NM_152458
|
ZNF785
|
zinc finger protein 785
|
|
chr1_+_67546143
|
0.662
|
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chr1_-_158161877
|
0.653
|
|
TAGLN2
|
transgelin 2
|
|
chr1_-_158161902
|
0.652
|
NM_003564
|
TAGLN2
|
transgelin 2
|
|
chr3_-_16530132
|
0.648
|
NM_015150
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr1_+_199975563
|
0.643
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr10_-_43390014
|
0.640
|
NM_001099282
NM_001099283
NM_001099284
|
ZNF239
|
zinc finger protein 239
|
|
chr12_+_129922589
|
0.640
|
|
RAN
|
RAN, member RAS oncogene family
|
|
chr16_-_53520208
|
0.638
|
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr5_-_43432969
|
0.637
|
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr8_+_49083522
|
0.628
|
NM_003350
|
UBE2V2
|
ubiquitin-conjugating enzyme E2 variant 2
|
|
chr4_-_2905649
|
0.625
|
NM_001120
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr5_+_49998677
|
0.623
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr16_-_80602546
|
0.623
|
NM_145168
|
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chrX_+_64804259
|
0.618
|
|
MSN
|
moesin
|
|
chr11_+_32808045
|
0.617
|
NM_024081
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr19_+_1022208
|
0.614
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr2_-_227371698
|
0.611
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr20_-_57015698
|
0.606
|
NM_001336
|
CTSZ
|
cathepsin Z
|
|
chrX_+_102356659
|
0.606
|
NM_001080425
NM_001127688
|
BEX4
|
brain expressed, X-linked 4
|
|
chr12_+_129922570
|
0.599
|
|
RAN
|
RAN, member RAS oncogene family
|
|
chr22_+_43527101
|
0.597
|
NM_001017526
NM_001198726
NM_181335
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr2_-_235070431
|
0.593
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr8_-_97342971
|
0.592
|
NM_015942
|
MTERFD1
|
MTERF domain containing 1
|
|
chr10_+_80672842
|
0.589
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr7_+_27102197
|
0.585
|
|
|
|
|
chr13_-_19633182
|
0.578
|
NM_021954
|
GJA3
|
gap junction protein, alpha 3, 46kDa
|
|
chr9_+_123501186
|
0.578
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr14_-_87529212
|
0.563
|
|
GALC
|
galactosylceramidase
|
|
chr8_-_75396016
|
0.561
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chrX_-_51256166
|
0.560
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr7_-_27102049
|
0.556
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr16_+_55180767
|
0.555
|
NM_005954
|
MT3
|
metallothionein 3
|
|
chr1_-_164402575
|
0.553
|
NM_001017961
|
FAM78B
|
family with sequence similarity 78, member B
|
|
chr3_-_69517913
|
0.552
|
|
FRMD4B
|
FERM domain containing 4B
|
|
chr14_-_87529222
|
0.548
|
|
GALC
|
galactosylceramidase
|
|
chr17_-_2561676
|
0.546
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr15_+_46957556
|
0.544
|
NM_014335
|
EID1
|
EP300 interacting inhibitor of differentiation 1
|
|
chr3_+_23219707
|
0.542
|
NM_152653
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast)
|
|
chr18_+_32131628
|
0.536
|
NM_025135
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr5_-_127447197
|
0.535
|
|
FLJ33630
|
hypothetical LOC644873
|
|
chr3_-_16529996
|
0.535
|
|
RFTN1
|
raftlin, lipid raft linker 1
|
|
chr13_-_35818491
|
0.534
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr17_-_10042579
|
0.534
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr9_+_111442888
|
0.531
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr11_+_65311104
|
0.531
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr17_+_44927591
|
0.528
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr21_-_41140908
|
0.524
|
NM_001389
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr22_+_43476755
|
0.523
|
NM_181334
|
PRR5-ARHGAP8
PRR5
|
PRR5-ARHGAP8 readthrough
proline rich 5 (renal)
|
|
chr9_+_92603884
|
0.520
|
|
SYK
|
spleen tyrosine kinase
|
|
chr13_-_35818396
|
0.518
|
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr4_+_78298326
|
0.516
|
|
CCNG2
|
cyclin G2
|
|
chrX_+_24077649
|
0.512
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr20_+_55718718
|
0.511
|
|
|
|
|
chr4_+_5104140
|
0.510
|
|
STK32B
|
serine/threonine kinase 32B
|
|
chr8_+_145718702
|
0.509
|
|
|
|
|
chrX_-_102452461
|
0.508
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr16_+_88305743
|
0.508
|
|
LOC100128881
|
hypothetical LOC100128881
|
|
chr4_-_2905448
|
0.507
|
|
MFSD10
|
major facilitator superfamily domain containing 10
|
|
chr1_+_199519202
|
0.501
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr14_-_87529600
|
0.498
|
NM_000153
NM_001037525
|
GALC
|
galactosylceramidase
|
|
chr22_-_16020153
|
0.497
|
|
CECR5
|
cat eye syndrome chromosome region, candidate 5
|
|
chr9_-_109291575
|
0.492
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr11_+_125279481
|
0.490
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr7_-_87687302
|
0.487
|
NM_003130
|
SRI
|
sorcin
|
|
chr19_-_36531958
|
0.483
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr3_-_69518119
|
0.481
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr1_+_182622987
|
0.481
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr11_-_63770716
|
0.479
|
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chr9_+_111582317
|
0.479
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr22_-_16020076
|
0.479
|
|
CECR5
|
cat eye syndrome chromosome region, candidate 5
|
|
chr5_+_176493488
|
0.478
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr1_+_45565126
|
0.477
|
NM_032756
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like
|
|
chr17_-_8000362
|
0.476
|
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr3_+_52694975
|
0.474
|
NM_014366
NM_206825
NM_206826
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr20_-_49073023
|
0.474
|
NM_002237
|
KCNG1
|
potassium voltage-gated channel, subfamily G, member 1
|
|
chr2_-_253982
|
0.472
|
|
SH3YL1
|
SH3 domain containing, Ysc84-like 1 (S. cerevisiae)
|
|
chr19_+_43502380
|
0.471
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr3_+_23219490
|
0.471
|
|
UBE2E2
|
ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast)
|
|
chr7_+_117611321
|
0.468
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr2_-_254037
|
0.465
|
NM_001159597
NM_015677
|
SH3YL1
|
SH3 domain containing, Ysc84-like 1 (S. cerevisiae)
|
|
chr3_+_120904555
|
0.465
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr19_-_60587414
|
0.461
|
NM_001190764
|
LOC388564
|
hypothetical protein LOC388564
|
|
chr6_+_72949092
|
0.460
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr2_-_119322228
|
0.459
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr19_+_40337846
|
0.459
|
|
FXYD5
|
FXYD domain containing ion transport regulator 5
|
|
chr3_+_114414064
|
0.458
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr1_+_181025494
|
0.457
|
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr8_-_72918938
|
0.453
|
|
MSC
|
musculin
|
|
chr22_-_41913002
|
0.450
|
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12
|
|
chr11_+_65311142
|
0.450
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr7_+_154720547
|
0.448
|
|
INSIG1
|
insulin induced gene 1
|
|
chr7_-_87687276
|
0.448
|
|
SRI
|
sorcin
|
|
chr1_-_173428455
|
0.448
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr4_-_122063118
|
0.447
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr22_+_48971067
|
0.445
|
|
TRABD
|
TraB domain containing
|
|
chr21_-_31853160
|
0.444
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr1_-_35957314
|
0.443
|
NM_152374
|
C1orf216
|
chromosome 1 open reading frame 216
|
|
chr8_-_9046460
|
0.442
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr1_+_117253882
|
0.439
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr9_+_92603894
|
0.439
|
|
SYK
|
spleen tyrosine kinase
|
|
chr10_+_105334704
|
0.437
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr7_+_148613304
|
0.435
|
|
LOC155060
|
AI894139 pseudogene
|
|
chr1_+_67545634
|
0.435
|
NM_001559
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chrX_+_64804318
|
0.434
|
|
MSN
|
moesin
|