|
chr2_-_188127294
|
1.539
|
|
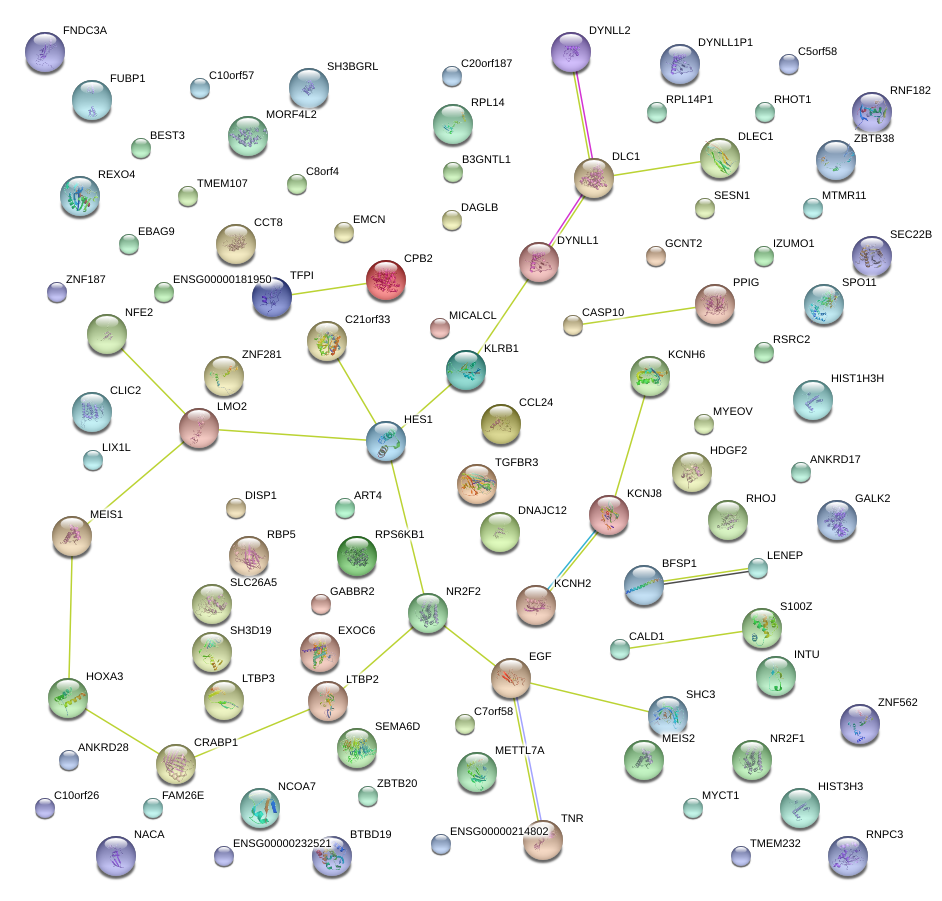
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr2_-_188127430
|
1.503
|
NM_001032281
NM_006287
|
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor)
|
|
chr13_+_48582389
|
1.475
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr10_+_94584449
|
1.412
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr12_-_14887648
|
1.336
|
NM_021071
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group)
|
|
chr5_-_110090264
|
1.185
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr7_+_120416680
|
0.884
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr8_+_40130143
|
0.882
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr11_+_68818175
|
0.875
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr17_-_59817456
|
0.871
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr17_-_57023322
|
0.864
|
NM_199290
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2
|
|
chr15_+_94670160
|
0.852
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr17_-_59817722
|
0.850
|
NM_000442
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr17_-_59817654
|
0.841
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr9_-_100510657
|
0.795
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr17_-_59817575
|
0.773
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr1_-_78217278
|
0.759
|
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr19_-_53941835
|
0.736
|
NM_182575
|
IZUMO1
|
izumo sperm-egg fusion 1
|
|
chr4_-_101658094
|
0.733
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr6_+_153060722
|
0.704
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chr1_+_45046740
|
0.703
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr20_+_55338221
|
0.670
|
NM_012444
NM_198265
|
SPO11
|
SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae)
|
|
chr12_-_121577410
|
0.652
|
NM_023012
|
RSRC2
|
arginine/serine-rich coiled-coil 2
|
|
chr4_-_152366970
|
0.644
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr8_-_13018123
|
0.643
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr14_+_62740878
|
0.642
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr10_+_104525877
|
0.639
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr12_+_49604782
|
0.635
|
NM_014033
|
METTL7A
|
methyltransferase like 7A
|
|
chrX_+_80344253
|
0.629
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chr11_+_65027721
|
0.621
|
|
|
|
|
chr15_+_47249700
|
0.618
|
NM_002044
|
GALK2
|
galactokinase 2
|
|
chr12_+_49605010
|
0.614
|
|
METTL7A
|
methyltransferase like 7A
|
|
chrX_-_102828238
|
0.602
|
NM_001142418
NM_001142423
NM_001142424
NM_001142425
NM_001142426
NM_001142429
NM_001142430
NM_012286
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr17_-_78599695
|
0.596
|
|
B3GNTL1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1
|
|
chr6_+_10663934
|
0.592
|
NM_001491
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr6_-_109521969
|
0.573
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chrX_+_80344215
|
0.572
|
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like
|
|
chrX_-_154216929
|
0.566
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr7_-_150283794
|
0.558
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr4_+_111053488
|
0.558
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr2_+_66516196
|
0.552
|
|
MEIS1
|
Meis homeobox 1
|
|
chr5_+_76181581
|
0.550
|
NM_130772
|
S100Z
|
S100 calcium binding protein Z
|
|
chr6_+_14032655
|
0.548
|
NM_001165034
|
RNF182
|
ring finger protein 182
|
|
chr1_+_202033273
|
0.535
|
NM_001174108
|
ZBED6
|
zinc finger, BED-type containing 6
|
|
chr10_-_69267774
|
0.523
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr3_+_142569988
|
0.521
|
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr20_-_17487480
|
0.519
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr7_-_6441109
|
0.500
|
|
DAGLB
|
diacylglycerol lipase, beta
|
|
chr12_-_52975823
|
0.495
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr6_-_109522207
|
0.491
|
|
SESN1
|
sestrin 1
|
|
chr1_+_221168388
|
0.490
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr4_-_74307637
|
0.486
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr19_+_4442610
|
0.472
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr1_+_143807876
|
0.470
|
|
SEC22B
|
SEC22 vesicle trafficking protein homolog B (S. cerevisiae) (gene/pseudogene)
|
|
chr3_-_115585178
|
0.469
|
NM_001164342
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr12_-_68369322
|
0.466
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr6_+_126143971
|
0.451
|
|
NCOA7
|
nuclear receptor coactivator 7
|
|
chr7_+_134114701
|
0.451
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr1_-_173979292
|
0.448
|
NM_003285
|
TNR
|
tenascin R (restrictin, janusin)
|
|
chr3_+_195336625
|
0.440
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr7_-_27133163
|
0.439
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr11_+_12265022
|
0.439
|
NM_032867
|
MICALCL
|
MICAL C-terminal like
|
|
chr15_+_45263517
|
0.439
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr3_-_15814678
|
0.436
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr12_-_21818881
|
0.433
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr1_+_153232685
|
0.431
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr4_+_128773529
|
0.414
|
NM_015693
|
INTU
|
inturned planar cell polarity effector homolog (Drosophila)
|
|
chr7_-_75280962
|
0.411
|
NM_002991
|
CCL24
|
chemokine (C-C motif) ligand 24
|
|
chr5_+_169592523
|
0.410
|
NM_001102609
|
C5orf58
|
chromosome 5 open reading frame 58
|
|
chr11_-_33870411
|
0.409
|
NM_005574
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr7_+_134114957
|
0.406
|
|
CALD1
|
caldesmon 1
|
|
chr13_-_45577144
|
0.401
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr6_+_116939500
|
0.393
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr1_-_148174722
|
0.392
|
NM_181873
|
MTMR11
|
myotubularin related protein 11
|
|
chr2_+_201756048
|
0.391
|
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr6_+_28342766
|
0.391
|
NM_001023560
NM_001111039
NM_152736
|
ZNF187
|
zinc finger protein 187
|
|
chr8_+_110621485
|
0.391
|
NM_004215
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9
|
|
chr1_-_226679648
|
0.388
|
NM_003493
|
HIST3H3
|
histone cluster 3, H3
|
|
chr1_-_92144146
|
0.388
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr10_+_82163568
|
0.383
|
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr1_-_198644111
|
0.382
|
|
ZNF281
|
zinc finger protein 281
|
|
chr1_+_109450116
|
0.378
|
|
|
|
|
chr7_-_102873747
|
0.377
|
NM_001167962
NM_198999
NM_206883
NM_206884
NM_206885
|
SLC26A5
|
solute carrier family 26, member 5 (prestin)
|
|
chr9_-_90983195
|
0.376
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr1_+_144188375
|
0.374
|
NM_153713
|
LIX1L
|
Lix1 homolog (mouse)-like
|
|
chr12_-_7172641
|
0.372
|
NM_031491
|
RBP5
|
retinol binding protein 5, cellular
|
|
chr17_+_27493680
|
0.372
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr11_-_65081967
|
0.372
|
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr12_+_61645361
|
0.370
|
|
RPL14
|
ribosomal protein L14
|
|
chr21_-_29367820
|
0.366
|
|
CCT8
|
chaperonin containing TCP1, subunit 8 (theta)
|
|
chr12_-_14611820
|
0.364
|
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr2_-_192419896
|
0.363
|
|
SDPR
|
serum deprivation response
|
|
chr11_+_66764060
|
0.362
|
|
LOC100131150
|
hypothetical protein LOC100131150
|
|
chr21_-_29367784
|
0.362
|
|
CCT8
|
chaperonin containing TCP1, subunit 8 (theta)
|
|
chr17_-_74361975
|
0.361
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr4_+_47403107
|
0.361
|
|
|
|
|
chr17_-_36915329
|
0.361
|
NM_002274
NM_153490
|
KRT13
|
keratin 13
|
|
chr3_+_52504410
|
0.357
|
|
STAB1
|
stabilin 1
|
|
chr6_+_32229278
|
0.354
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr13_+_96672541
|
0.354
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr2_-_113259441
|
0.353
|
NM_000575
|
IL1A
|
interleukin 1, alpha
|
|
chr21_-_29367839
|
0.351
|
NM_006585
|
CCT8
|
chaperonin containing TCP1, subunit 8 (theta)
|
|
chr6_+_27323458
|
0.351
|
NM_005865
|
PRSS16
|
protease, serine, 16 (thymus)
|
|
chr9_-_74757735
|
0.348
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr6_+_112775224
|
0.347
|
NM_001013734
|
RFPL4B
|
ret finger protein-like 4B
|
|
chr2_-_136592099
|
0.344
|
NM_003467
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr10_+_70517863
|
0.342
|
|
SRGN
|
serglycin
|
|
chr2_+_128119908
|
0.341
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr3_+_46514488
|
0.341
|
NM_031440
|
RTP3
|
receptor (chemosensory) transporter protein 3
|
|
chr22_+_19458378
|
0.339
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr8_+_39890484
|
0.337
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr20_+_29598848
|
0.337
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr5_+_171553780
|
0.336
|
NM_001171183
|
EFCAB9
|
EF-hand calcium binding domain 9
|
|
chr11_+_28088406
|
0.336
|
|
METTL15
|
methyltransferase like 15
|
|
chr18_-_50776276
|
0.335
|
NM_001143829
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr5_+_67622242
|
0.335
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_-_78217360
|
0.334
|
NM_003902
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr3_+_45705845
|
0.334
|
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast)
|
|
chr19_-_14490160
|
0.331
|
NM_006145
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr10_+_70517823
|
0.331
|
NM_002727
|
SRGN
|
serglycin
|
|
chr7_+_134114912
|
0.330
|
|
CALD1
|
caldesmon 1
|
|
chr13_+_96672705
|
0.328
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chrX_-_80344096
|
0.328
|
NM_030763
|
HMGN5
|
high-mobility group nucleosome binding domain 5
|
|
chr12_+_50633430
|
0.327
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr14_-_22458199
|
0.326
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr18_-_50777634
|
0.324
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr14_-_22610572
|
0.323
|
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chrX_+_47999740
|
0.322
|
NM_005635
|
SSX1
|
synovial sarcoma, X breakpoint 1
|
|
chr11_+_66764064
|
0.320
|
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr12_-_14612057
|
0.319
|
NM_024829
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr19_-_14121728
|
0.318
|
|
LPHN1
|
latrophilin 1
|
|
chr13_-_40454188
|
0.318
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr1_-_180621272
|
0.317
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr12_+_118100977
|
0.316
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr8_-_110689396
|
0.314
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr6_-_111995213
|
0.314
|
NM_001164283
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr3_-_124893875
|
0.313
|
|
MYLK
|
myosin light chain kinase
|
|
chr14_-_22458220
|
0.311
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr3_+_45705918
|
0.311
|
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast)
|
|
chr4_+_91035025
|
0.311
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr3_-_184756101
|
0.311
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr12_-_69317441
|
0.311
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr15_+_40481879
|
0.309
|
NM_173088
|
CAPN3
|
calpain 3, (p94)
|
|
chr1_-_148175345
|
0.308
|
|
MTMR11
|
myotubularin related protein 11
|
|
chr1_+_154095883
|
0.308
|
NM_152280
|
SYT11
|
synaptotagmin XI
|
|
chr2_+_26768894
|
0.307
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr12_-_52975777
|
0.307
|
NM_006163
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr11_+_65526462
|
0.304
|
|
BANF1
|
barrier to autointegration factor 1
|
|
chr14_-_22458134
|
0.304
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr2_+_201755865
|
0.304
|
NM_001230
NM_032974
NM_032977
|
CASP10
|
caspase 10, apoptosis-related cysteine peptidase
|
|
chr3_-_19950675
|
0.303
|
NM_144715
|
EFHB
|
EF-hand domain family, member B
|
|
chr1_+_15858950
|
0.301
|
NM_006511
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1
|
|
chr6_-_161614962
|
0.301
|
NM_020133
|
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr3_+_69871252
|
0.301
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_-_93181091
|
0.300
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr15_-_54175954
|
0.300
|
|
RFX7
|
regulatory factor X, 7
|
|
chr3_+_52504391
|
0.299
|
NM_015136
|
STAB1
|
stabilin 1
|
|
chr3_+_45705681
|
0.299
|
NM_014016
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast)
|
|
chr13_-_40735619
|
0.297
|
NM_004294
|
MTRF1
|
mitochondrial translational release factor 1
|
|
chr10_+_74540215
|
0.297
|
NM_015901
|
NUDT13
|
nudix (nucleoside diphosphate linked moiety X)-type motif 13
|
|
chrX_-_102829595
|
0.297
|
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr7_+_134483305
|
0.296
|
NM_018295
|
TMEM140
|
transmembrane protein 140
|
|
chr16_+_4842077
|
0.296
|
NM_016936
|
UBN1
|
ubinuclein 1
|
|
chr1_+_114295289
|
0.294
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr17_-_74361620
|
0.294
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr17_+_27493654
|
0.289
|
|
RHOT1
|
ras homolog gene family, member T1
|
|
chr2_+_160180092
|
0.289
|
|
LOC643072
|
hypothetical LOC643072
|
|
chr10_+_18280773
|
0.287
|
NM_001145195
NM_152725
|
SLC39A12
|
solute carrier family 39 (zinc transporter), member 12
|
|
chr6_-_76010231
|
0.287
|
NM_001865
|
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver)
|
|
chr13_-_40454360
|
0.287
|
NM_001145353
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr8_-_31010772
|
0.287
|
NM_001015508
|
PURG
|
purine-rich element binding protein G
|
|
chr1_-_232810111
|
0.286
|
|
IRF2BP2
|
interferon regulatory factor 2 binding protein 2
|
|
chr6_-_68655708
|
0.286
|
|
|
|
|
chr7_+_18296569
|
0.286
|
|
HDAC9
|
histone deacetylase 9
|
|
chr6_-_135417522
|
0.286
|
|
HBS1L
|
HBS1-like (S. cerevisiae)
|
|
chr2_-_158439868
|
0.286
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chrX_+_48529905
|
0.282
|
NM_002049
|
GATA1
|
GATA binding protein 1 (globin transcription factor 1)
|
|
chr19_+_13066956
|
0.281
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr2_-_54336847
|
0.279
|
NM_001003937
|
TSPYL6
|
TSPY-like 6
|
|
chr3_+_195336631
|
0.272
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr2_+_160180049
|
0.271
|
|
LOC643072
|
hypothetical LOC643072
|
|
chr3_-_71262405
|
0.267
|
|
FOXP1
|
forkhead box P1
|
|
chr8_-_82769761
|
0.266
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr7_+_105304445
|
0.266
|
|
LOC100506768
CDHR3
|
hypothetical LOC100506768
cadherin-related family member 3
|
|
chr14_-_22610610
|
0.266
|
NM_001164816
NM_001164817
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr3_-_140677990
|
0.265
|
NM_004164
|
RBP2
|
retinol binding protein 2, cellular
|
|
chr1_-_94147537
|
0.265
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr17_-_44058612
|
0.264
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr5_-_180175146
|
0.264
|
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr21_-_30720100
|
0.263
|
NM_181622
|
KRTAP13-3
|
keratin associated protein 13-3
|
|
chr1_-_196011152
|
0.261
|
NM_001195215
NM_001195216
NM_144977
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr1_-_171442937
|
0.260
|
NM_003326
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr6_+_88026338
|
0.259
|
|
ZNF292
|
zinc finger protein 292
|
|
chr19_+_50041424
|
0.256
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr7_-_27125738
|
0.254
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr1_+_66570683
|
0.253
|
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr6_+_32230172
|
0.253
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr8_+_31010791
|
0.251
|
|
WRN
|
Werner syndrome, RecQ helicase-like
|
|
chrX_-_131181141
|
0.251
|
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr6_+_167624792
|
0.251
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|