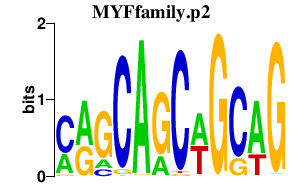
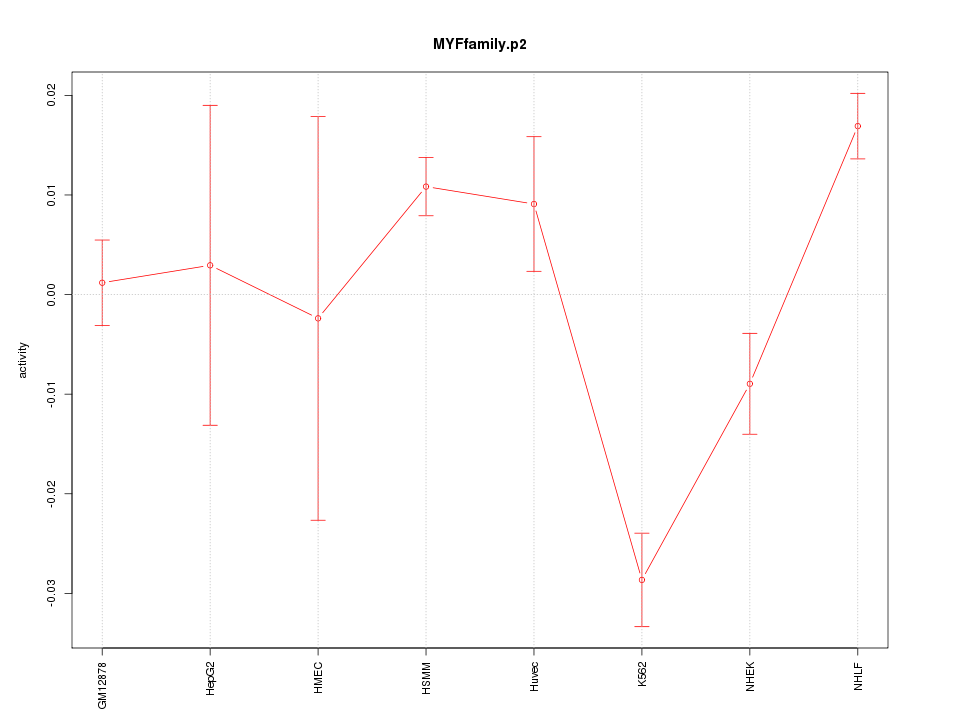
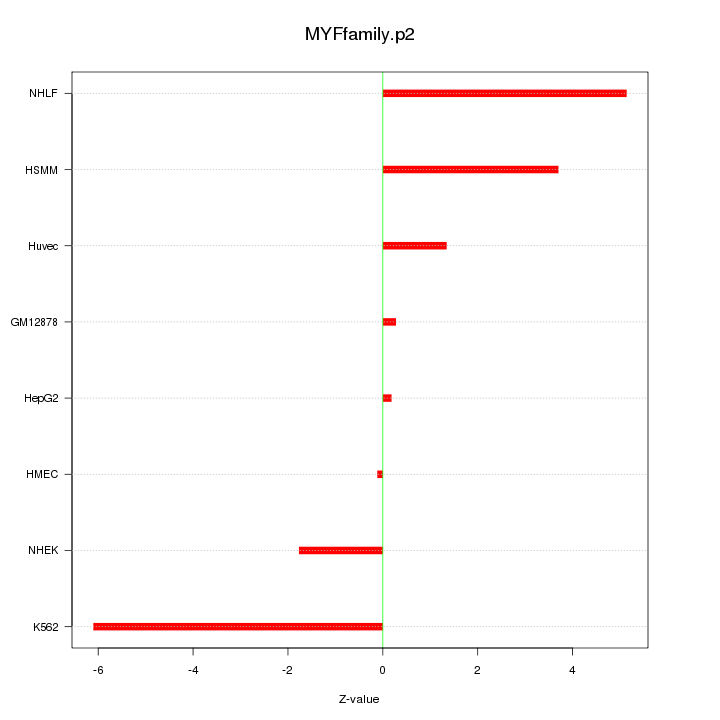
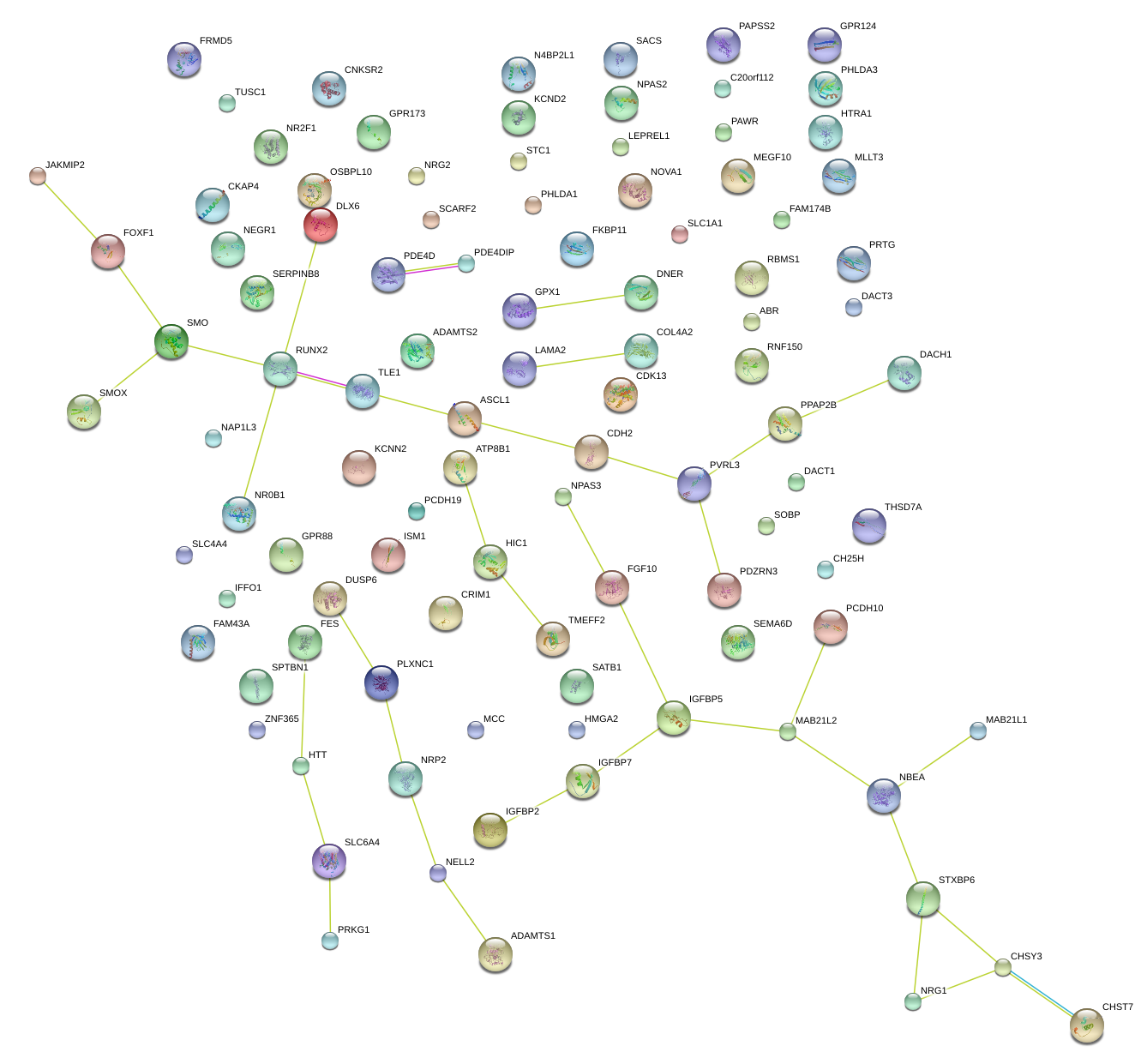
|
chr5_-_139402909
|
7.152
|
|
NRG2
|
neuregulin 2
|
|
chr5_-_139402989
|
6.118
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chr13_-_71338558
|
5.736
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr4_+_3046034
|
5.637
|
NM_002111
|
HTT
|
huntingtin
|
|
chr8_-_23768242
|
5.542
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr3_-_31997738
|
5.046
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr13_-_71338144
|
4.994
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr12_-_74710741
|
4.953
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr13_+_34948885
|
4.685
|
|
NBEA
|
neurobeachin
|
|
chr13_-_34948758
|
4.670
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr18_-_24010944
|
4.012
|
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr18_+_59788242
|
3.751
|
NM_001031848
NM_002640
NM_198833
|
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8
|
|
chr2_-_230287442
|
3.615
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr6_+_107917965
|
3.546
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr12_+_64504835
|
3.441
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr9_+_4480193
|
3.336
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr9_-_83494198
|
3.326
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr5_-_112852425
|
3.324
|
NM_001085377
|
MCC
|
mutated in colorectal cancers
|
|
chr10_+_89409332
|
3.286
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr14_-_26136799
|
3.286
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr1_-_72521079
|
3.237
|
|
|
|
|
chr1_+_100777044
|
3.186
|
|
GPR88
|
G protein-coupled receptor 88
|
|
chr12_-_6535481
|
3.132
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr10_-_90957028
|
3.097
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr6_+_129245978
|
3.035
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr5_+_113725914
|
3.035
|
NM_021614
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr12_-_88269884
|
2.965
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr20_-_30534848
|
2.937
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr4_+_159311353
|
2.933
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr20_-_30535045
|
2.933
|
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr5_-_178705036
|
2.906
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr1_-_56817802
|
2.836
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr22_-_19122047
|
2.831
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr3_-_191321601
|
2.661
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr3_+_195889845
|
2.596
|
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr12_-_88270128
|
2.593
|
|
DUSP6
|
dual specificity phosphatase 6
|
|
chr2_-_192767844
|
2.578
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr5_+_92946348
|
2.555
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr4_-_142273881
|
2.503
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr6_+_45498354
|
2.501
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chrX_-_92815222
|
2.499
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr1_-_199704773
|
2.476
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr12_+_93066370
|
2.461
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr5_-_178704934
|
2.413
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr1_-_72520735
|
2.392
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr17_-_959007
|
2.359
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr7_+_128615948
|
2.310
|
NM_005631
|
SMO
|
smoothened homolog (Drosophila)
|
|
chrX_-_99551926
|
2.280
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chrX_+_53095230
|
2.271
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr1_-_56817568
|
2.230
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr16_+_85101734
|
2.216
|
|
FOXF1
|
forkhead box F1
|
|
chr8_+_37773991
|
2.215
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr18_-_53621277
|
2.201
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr16_+_85101605
|
2.189
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr9_-_25668230
|
2.171
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr14_-_24588916
|
2.167
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr2_+_36436873
|
2.161
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr5_+_113726203
|
2.161
|
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chrX_+_21302317
|
2.074
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chrX_+_46318135
|
2.059
|
NM_019886
|
CHST7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7
|
|
chr1_-_56817717
|
2.041
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_+_129268352
|
2.038
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr15_+_45796802
|
2.034
|
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr8_+_31616809
|
2.032
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr17_+_1906262
|
2.016
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr15_-_90999606
|
1.996
|
NM_207446
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr5_+_126654354
|
1.984
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr5_-_44424423
|
1.972
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chrX_-_30237403
|
1.966
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr10_+_52504239
|
1.958
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr13_-_22905822
|
1.915
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr13_-_31900218
|
1.915
|
NM_001079691
NM_052818
|
N4BP2L1
|
NEDD4 binding protein 2-like 1
|
|
chr12_+_101876151
|
1.908
|
|
ASCL1
|
achaete-scute complex homolog 1 (Drosophila)
|
|
chr12_-_105165805
|
1.905
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr7_+_39956633
|
1.889
|
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr15_-_53822468
|
1.885
|
NM_173814
|
PRTG
|
protogenin
|
|
chr5_-_59225377
|
1.881
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_18441763
|
1.869
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr15_+_89229275
|
1.827
|
NM_001143783
NM_001143784
|
FES
|
feline sarcoma oncogene
|
|
chr14_+_58174489
|
1.808
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr13_+_109757593
|
1.802
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr14_+_32478209
|
1.802
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr10_+_63803921
|
1.790
|
NM_014951
NM_199450
NM_199451
|
ZNF365
|
zinc finger protein 365
|
|
chr1_-_143786993
|
1.786
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_+_217206450
|
1.778
|
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr12_-_47605478
|
1.772
|
NM_001143782
NM_016594
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr18_-_24011349
|
1.768
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr3_-_49370682
|
1.760
|
|
GPX1
|
glutathione peroxidase 1
|
|
chr3_+_112273464
|
1.756
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr4_+_72271218
|
1.754
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr10_+_124210980
|
1.747
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr5_-_147142457
|
1.747
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr12_-_74711474
|
1.743
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr4_-_57671307
|
1.743
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr12_-_78608154
|
1.740
|
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr6_+_45498262
|
1.733
|
|
RUNX2
|
runt-related transcription factor 2
|
|
chr7_+_96473225
|
1.729
|
NM_005222
|
DLX6
|
distal-less homeobox 6
|
|
chr20_+_13150417
|
1.693
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr21_-_27139143
|
1.682
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr4_-_57671273
|
1.676
|
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr3_-_73756668
|
1.670
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr15_-_42274529
|
1.669
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr4_-_57671112
|
1.656
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr2_+_54536780
|
1.651
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr9_-_20612476
|
1.647
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr2_+_100802923
|
1.647
|
NM_002518
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr3_+_112273352
|
1.637
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr2_-_217267742
|
1.624
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr2_+_206255597
|
1.620
|
|
NRP2
|
neuropilin 2
|
|
chr7_+_119700924
|
1.618
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr2_-_161058120
|
1.587
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr7_-_11838260
|
1.586
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr2_+_191454036
|
1.577
|
|
GLS
|
glutaminase
|
|
chr9_+_35896188
|
1.569
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr7_-_84654106
|
1.558
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr3_+_141136896
|
1.540
|
|
CLSTN2
|
calsyntenin 2
|
|
chr3_+_185762233
|
1.519
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr9_+_108665168
|
1.507
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr13_+_34414390
|
1.504
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr8_-_89408832
|
1.487
|
NM_005941
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted)
|
|
chr14_-_59407103
|
1.484
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr6_+_127481496
|
1.483
|
|
RSPO3
|
R-spondin 3 homolog (Xenopus laevis)
|
|
chr8_+_37773915
|
1.477
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr9_-_129656413
|
1.458
|
|
ENG
|
endoglin
|
|
chr9_-_129656581
|
1.456
|
|
ENG
|
endoglin
|
|
chrX_-_154341451
|
1.455
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr3_-_87122936
|
1.444
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr2_-_216944911
|
1.435
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr5_-_147142444
|
1.423
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr1_-_59020875
|
1.411
|
|
JUN
|
jun proto-oncogene
|
|
chr7_-_137182125
|
1.408
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr11_-_129803712
|
1.390
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr9_-_128924706
|
1.388
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr6_+_1556543
|
1.388
|
|
FOXC1
|
forkhead box C1
|
|
chr3_-_79151232
|
1.387
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr4_+_37569093
|
1.369
|
NM_015173
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr2_-_160362914
|
1.365
|
|
CD302
|
CD302 molecule
|
|
chr3_-_87122958
|
1.354
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr4_-_96689373
|
1.348
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr2_-_227371698
|
1.344
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_+_126579203
|
1.342
|
NM_182487
|
OLFML2A
|
olfactomedin-like 2A
|
|
chr2_-_160362982
|
1.341
|
NM_001198763
NM_001198764
NM_014880
|
CD302
|
CD302 molecule
|
|
chr10_+_90629880
|
1.335
|
NM_020799
|
STAMBPL1
|
STAM binding protein-like 1
|
|
chr4_+_15313850
|
1.335
|
|
BST1
|
bone marrow stromal cell antigen 1
|
|
chr6_+_30002084
|
1.334
|
|
|
|
|
chr2_+_217206327
|
1.334
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr9_-_128924782
|
1.332
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr4_-_90448157
|
1.326
|
NM_198281
|
GPRIN3
|
GPRIN family member 3
|
|
chr2_-_227372574
|
1.325
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr15_+_79213660
|
1.325
|
NM_173528
|
C15orf26
|
chromosome 15 open reading frame 26
|
|
chrX_+_75564449
|
1.322
|
NM_020932
|
MAGEE1
|
melanoma antigen family E, 1
|
|
chr10_+_120957084
|
1.321
|
NM_005308
|
GRK5
|
G protein-coupled receptor kinase 5
|
|
chr6_+_114285209
|
1.320
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr4_+_134292630
|
1.319
|
|
PCDH10
|
protocadherin 10
|
|
chrX_-_106846253
|
1.314
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr15_+_89228668
|
1.312
|
NM_001143785
NM_002005
|
FES
|
feline sarcoma oncogene
|
|
chr2_-_75280121
|
1.308
|
NM_001058
NM_015727
|
TACR1
|
tachykinin receptor 1
|
|
chr15_+_97462675
|
1.306
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr7_+_68701704
|
1.298
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr9_-_129656778
|
1.295
|
|
ENG
|
endoglin
|
|
chr19_-_18912112
|
1.293
|
NM_001145722
NM_001145721
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr5_-_139992621
|
1.292
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr15_-_42274760
|
1.280
|
|
FRMD5
|
FERM domain containing 5
|
|
chr13_-_43259032
|
1.274
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr22_+_40286957
|
1.271
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr9_+_35782405
|
1.267
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr2_-_160627181
|
1.263
|
|
PLA2R1
|
phospholipase A2 receptor 1, 180kDa
|
|
chr13_-_36392370
|
1.263
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr12_-_54387732
|
1.262
|
|
ITGA7
|
integrin, alpha 7
|
|
chr1_+_144273662
|
1.257
|
|
ANKRD35
|
ankyrin repeat domain 35
|
|
chr4_+_169989680
|
1.251
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr9_+_27099138
|
1.248
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr3_-_147361626
|
1.246
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr3_+_160964574
|
1.243
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_-_29321599
|
1.228
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr2_+_207016525
|
1.222
|
NM_003812
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr22_+_44277354
|
1.213
|
NM_001996
NM_006485
NM_006486
NM_006487
|
FBLN1
|
fibulin 1
|
|
chr17_+_45488330
|
1.212
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr15_-_46724360
|
1.212
|
|
FBN1
|
fibrillin 1
|
|
chr13_+_34948977
|
1.211
|
|
NBEA
|
neurobeachin
|
|
chr2_+_36436316
|
1.210
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr6_+_127481740
|
1.210
|
NM_032784
|
RSPO3
|
R-spondin 3 homolog (Xenopus laevis)
|
|
chr2_-_161058550
|
1.206
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr9_-_90983195
|
1.206
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr12_-_54387934
|
1.202
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr4_+_109072120
|
1.200
|
NM_183075
|
CYP2U1
|
cytochrome P450, family 2, subfamily U, polypeptide 1
|
|
chr1_-_151852128
|
1.199
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr7_-_27190882
|
1.199
|
|
HOXA11
|
homeobox A11
|
|
chr15_+_45797977
|
1.198
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr2_+_187266899
|
1.197
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr5_-_147142368
|
1.196
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr3_-_191321367
|
1.196
|
|
LEPREL1
|
leprecan-like 1
|
|
chr22_-_19122107
|
1.190
|
NM_153334
NM_182895
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr3_-_147361465
|
1.189
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr9_-_129656830
|
1.187
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr14_-_51605487
|
1.179
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chrX_-_153767625
|
1.177
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chrX_+_153767803
|
1.175
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr9_-_130980302
|
1.175
|
|
IER5L
|
immediate early response 5-like
|
|
chr1_-_231498081
|
1.169
|
NM_014801
|
PCNXL2
|
pecanex-like 2 (Drosophila)
|