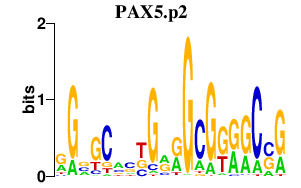
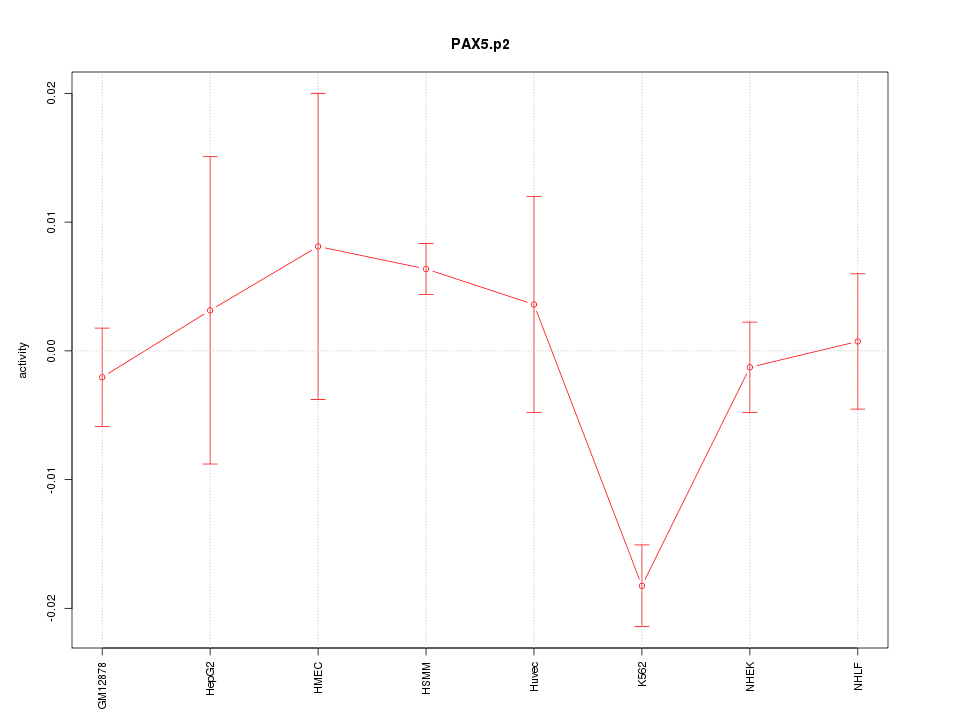
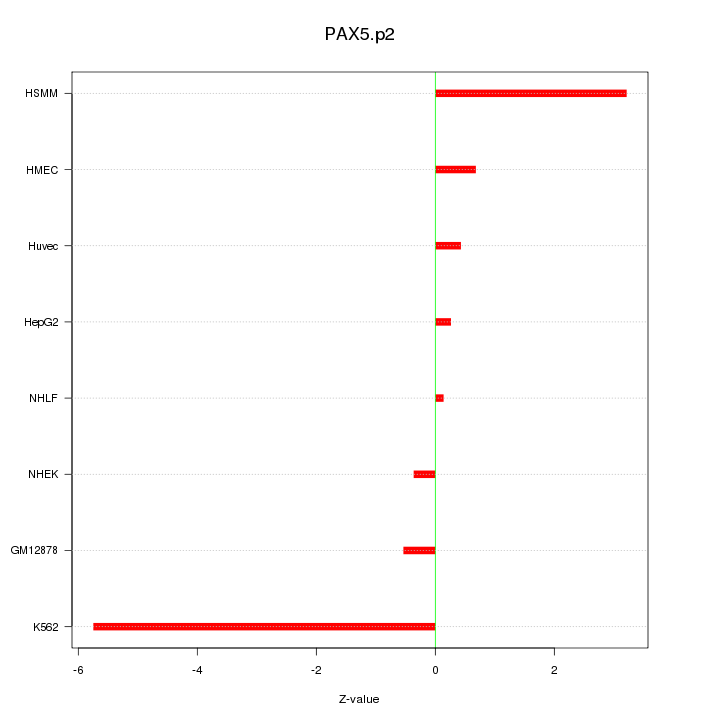
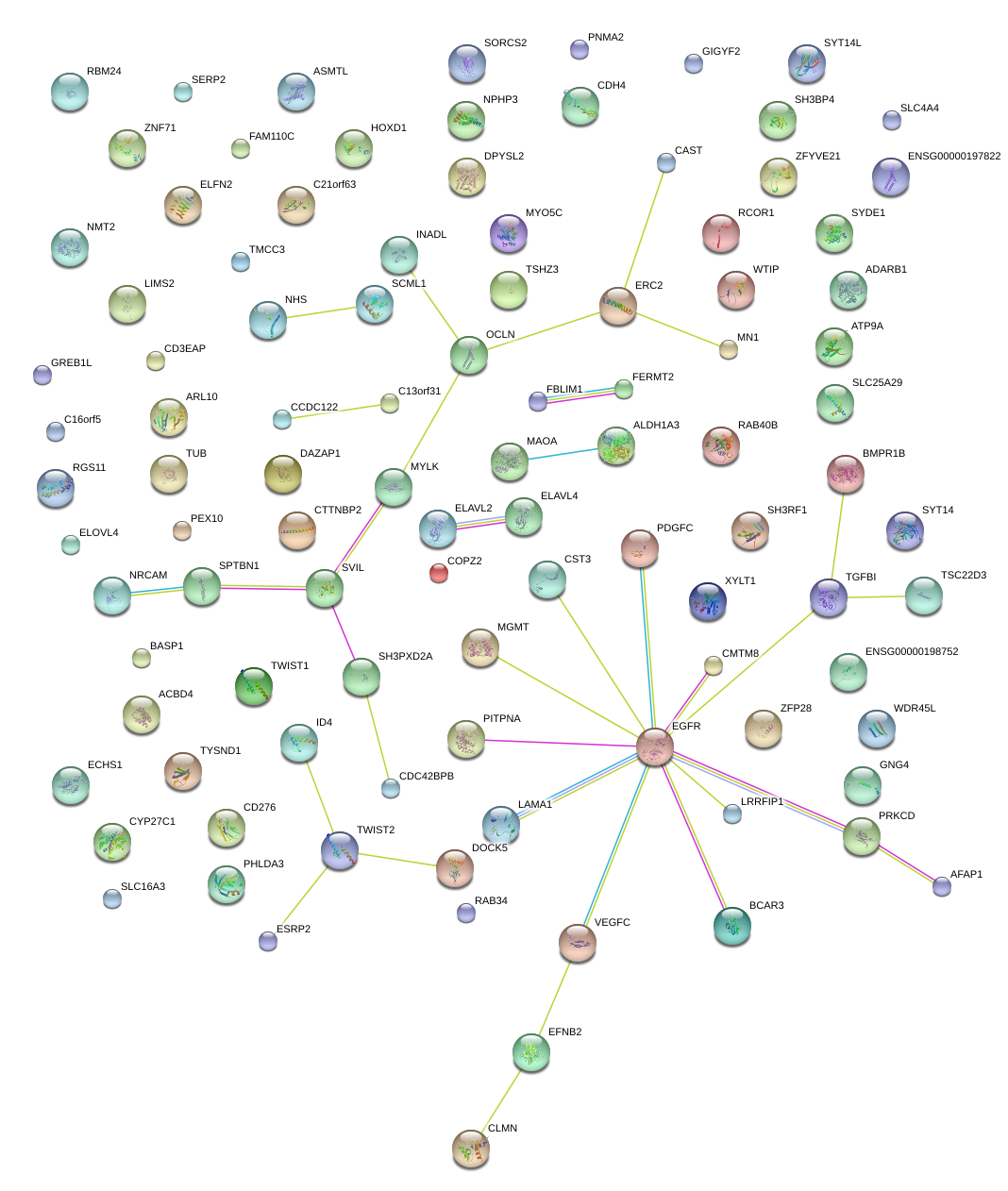
|
chr19_+_61742111
|
5.914
|
NM_020828
|
ZFP28
|
zinc finger protein 28 homolog (mouse)
|
|
chr1_-_233879617
|
5.342
|
NM_001098722
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr13_-_43351825
|
3.696
|
NM_144974
|
CCDC122
|
coiled-coil domain containing 122
|
|
chr17_-_78199384
|
3.436
|
NM_019613
|
WDR45L
|
WDR45-like
|
|
chr10_-_105604942
|
3.407
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr4_-_7991992
|
3.115
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr13_+_43351968
|
2.964
|
NM_153218
|
C13orf31
|
chromosome 13 open reading frame 31
|
|
chr12_-_93568336
|
2.947
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr10_-_30064600
|
2.904
|
NM_003174
|
SVIL
|
supervillin
|
|
chr5_+_96023926
|
2.862
|
|
CAST
|
calpastatin
|
|
chr2_+_235525355
|
2.825
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr2_-_36384
|
2.795
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr3_-_133923906
|
2.738
|
NM_153240
|
NPHP3
|
nephronophthisis 3 (adolescent)
|
|
chr7_-_107883929
|
2.693
|
NM_001193582
NM_001193583
NM_001193584
NM_005010
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr3_+_133923758
|
2.621
|
|
NCRNA00119
|
non-protein coding RNA 119
|
|
chr2_-_127694123
|
2.479
|
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1
|
|
chr3_-_133923992
|
2.470
|
|
NPHP3-ACAD11
|
NPHP3-ACAD11 read-through transcript
|
|
chr7_+_55054416
|
2.293
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr7_+_55054152
|
2.269
|
NM_005228
NM_201282
NM_201283
NM_201284
|
EGFR
|
epidermal growth factor receptor
|
|
chr5_+_17270781
|
2.246
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr1_+_15957832
|
2.164
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr8_+_25097983
|
2.115
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr16_-_66827449
|
2.114
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr19_-_36531958
|
2.111
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr13_+_43845977
|
2.031
|
NM_001010897
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2
|
|
chr3_+_32255158
|
1.946
|
NM_178868
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr14_+_102128844
|
1.922
|
NM_015156
|
RCOR1
|
REST corepressor 1
|
|
chr1_+_61980679
|
1.892
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr6_+_17389752
|
1.867
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr2_-_128149112
|
1.827
|
NM_001136037
NM_001161403
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr1_-_199704773
|
1.816
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr10_+_131155490
|
1.800
|
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr15_-_50375143
|
1.783
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr10_+_131155443
|
1.773
|
NM_002412
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr19_+_61798434
|
1.765
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr8_+_25098296
|
1.749
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr1_+_208178198
|
1.740
|
|
SYT14
|
synaptotagmin XIV
|
|
chr21_+_32706615
|
1.729
|
NM_058187
|
C21orf63
|
chromosome 21 open reading frame 63
|
|
chr15_+_99237527
|
1.727
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr19_+_39664706
|
1.726
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr5_+_17270474
|
1.721
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chrX_-_1531733
|
1.711
|
NM_001173474
NM_004192
|
ASMTL
|
acetylserotonin O-methyltransferase-like
|
|
chr11_+_8059267
|
1.680
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr4_-_177950658
|
1.675
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr21_+_45318861
|
1.669
|
NM_001112
NM_001160230
NM_015833
NM_015834
|
ADARB1
|
adenosine deaminase, RNA-specific, B1
|
|
chr4_-_170428681
|
1.639
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr4_-_170428564
|
1.632
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr17_+_40565492
|
1.630
|
NM_001135704
|
ACBD4
|
acyl-CoA binding domain containing 4
|
|
chr13_-_105985313
|
1.629
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr5_+_175725050
|
1.626
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr16_-_265868
|
1.624
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chr8_+_25098171
|
1.595
|
NM_024940
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr16_-_4528380
|
1.590
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr14_+_102128741
|
1.581
|
|
RCOR1
|
REST corepressor 1
|
|
chr10_-_71575953
|
1.576
|
|
TYSND1
|
trypsin domain containing 1
|
|
chr22_-_36153449
|
1.570
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr18_-_7107757
|
1.565
|
NM_005559
|
LAMA1
|
laminin, alpha 1
|
|
chr2_+_233270275
|
1.564
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr5_+_135392482
|
1.538
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr6_-_80713589
|
1.532
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr14_-_94855848
|
1.529
|
NM_024734
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr3_+_53170254
|
1.528
|
NM_006254
NM_212539
|
PRKCD
|
protein kinase C, delta
|
|
chr8_-_26427304
|
1.520
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr5_+_167889123
|
1.518
|
|
|
|
|
chr2_+_233270241
|
1.514
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr3_+_32255215
|
1.503
|
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr17_-_24068759
|
1.502
|
NM_001144942
NM_031934
|
RAB34
|
RAB34, member RAS oncogene family
|
|
chr7_-_19123787
|
1.487
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr4_+_7245218
|
1.485
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr19_+_15079141
|
1.474
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chrX_+_17665483
|
1.472
|
NM_001037535
NM_001037536
NM_001037540
NM_006746
|
SCML1
|
sex comb on midleg-like 1 (Drosophila)
|
|
chr5_+_68824143
|
1.467
|
|
OCLN
|
occludin
|
|
chrX_+_43399101
|
1.460
|
|
MAOA
|
monoamine oxidase A
|
|
chr4_+_72271218
|
1.440
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr17_-_78249750
|
1.436
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr4_+_95898098
|
1.428
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr10_-_135036805
|
1.425
|
NM_004092
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial
|
|
chr13_+_43351419
|
1.423
|
NM_001128303
|
C13orf31
|
chromosome 13 open reading frame 31
|
|
chr9_-_23811808
|
1.421
|
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr3_-_125085834
|
1.420
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr7_-_19123765
|
1.420
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr14_-_102593384
|
1.411
|
NM_006035
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like)
|
|
chr2_+_238265617
|
1.410
|
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr20_-_23566325
|
1.400
|
NM_000099
|
CST3
|
cystatin C
|
|
chr20_-_49818306
|
1.390
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr5_+_96023627
|
1.384
|
NM_001042440
|
CAST
|
calpastatin
|
|
chr7_-_117300763
|
1.382
|
NM_033427
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr17_-_1412774
|
1.381
|
NM_006224
|
PITPNA
|
phosphatidylinositol transfer protein, alpha
|
|
chr14_-_52487386
|
1.380
|
|
FERMT2
|
fermitin family member 2
|
|
chr16_+_65018316
|
1.370
|
NM_001136106
NM_001178020
NM_001197224
NM_001197225
|
BEAN1
|
brain expressed, associated with NEDD4, 1
|
|
chr1_-_93919789
|
1.370
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr4_-_158111995
|
1.367
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr12_+_13044604
|
1.361
|
|
HTR7P1
|
5-hydroxytryptamine (serotonin) receptor 7 pseudogene 1
|
|
chr22_-_26527469
|
1.360
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr2_+_238265519
|
1.359
|
NM_001137551
NM_001137552
NM_001137553
NM_004735
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr1_-_2333800
|
1.358
|
|
PEX10
|
peroxisomal biogenesis factor 10
|
|
chr8_+_26427378
|
1.354
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr5_+_96024434
|
1.349
|
|
CAST
|
calpastatin
|
|
chr15_+_71763830
|
1.340
|
|
CD276
|
CD276 molecule
|
|
chr2_+_54536780
|
1.340
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr15_+_71763742
|
1.335
|
|
CD276
|
CD276 molecule
|
|
chr16_-_17472238
|
1.334
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr6_+_19945578
|
1.326
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr20_+_59260847
|
1.309
|
NM_001794
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal)
|
|
chr14_+_103251798
|
1.302
|
NM_001198953
NM_024071
|
ZFYVE21
|
zinc finger, FYVE domain containing 21
|
|
chr20_-_49818268
|
1.299
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr1_+_208178160
|
1.288
|
NM_001146261
NM_001146262
NM_001146264
NM_153262
|
SYT14
|
synaptotagmin XIV
|
|
chr18_+_17076166
|
1.287
|
NM_001142966
|
GREB1L
|
growth regulation by estrogen in breast cancer-like
|
|
chr14_-_99842519
|
1.281
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr17_-_43470134
|
1.281
|
NM_016429
|
COPZ2
|
coatomer protein complex, subunit zeta 2
|
|
chr1_-_233880676
|
1.280
|
NM_001098721
NM_004485
|
GNG4
|
guanine nucleotide binding protein (G protein), gamma 4
|
|
chr10_-_15250616
|
1.273
|
NM_004808
|
NMT2
|
N-myristoyltransferase 2
|
|
chr17_+_77780177
|
1.257
|
NM_004207
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr15_+_71763773
|
1.253
|
|
CD276
|
CD276 molecule
|
|
chrX_-_106846253
|
1.246
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr15_+_71763674
|
1.237
|
NM_001024736
NM_025240
|
CD276
|
CD276 molecule
|
|
chr2_+_176761552
|
1.235
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr1_+_9275542
|
1.213
|
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr6_-_89884324
|
1.208
|
NM_080743
|
SRSF12
|
serine/arginine-rich splicing factor 12
|
|
chr1_-_2333863
|
1.207
|
NM_002617
NM_153818
|
PEX10
|
peroxisomal biogenesis factor 10
|
|
chr14_-_38971370
|
1.199
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr14_-_52487558
|
1.198
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr1_-_48235148
|
1.198
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr1_-_223907283
|
1.194
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr12_+_131000455
|
1.194
|
NM_015409
|
EP400
|
E1A binding protein p400
|
|
chr10_+_129595349
|
1.183
|
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr10_-_131652364
|
1.182
|
|
EBF3
|
early B-cell factor 3
|
|
chr3_-_38046034
|
1.172
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr4_+_183302105
|
1.171
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr4_-_170428768
|
1.162
|
NM_020870
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr6_-_56816403
|
1.162
|
|
DST
|
dystonin
|
|
chr20_+_13924014
|
1.156
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr8_-_125809559
|
1.154
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr12_+_50686990
|
1.151
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr17_-_25112393
|
1.147
|
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr6_+_83130673
|
1.144
|
NM_001166392
|
TPBG
|
trophoblast glycoprotein
|
|
chr19_-_1518875
|
1.143
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr13_+_34414390
|
1.142
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr16_-_87450734
|
1.140
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr3_+_128874458
|
1.132
|
NM_172027
|
ABTB1
|
ankyrin repeat and BTB (POZ) domain containing 1
|
|
chr7_+_288051
|
1.127
|
NM_020223
|
FAM20C
|
family with sequence similarity 20, member C
|
|
chr7_-_38637475
|
1.122
|
|
AMPH
|
amphiphysin
|
|
chr2_-_45090025
|
1.117
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr19_-_47619992
|
1.112
|
|
LIPE
|
lipase, hormone-sensitive
|
|
chr9_-_83494198
|
1.110
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr22_-_49292886
|
1.104
|
|
LMF2
|
lipase maturation factor 2
|
|
chr12_-_13044467
|
1.103
|
NM_015987
|
HEBP1
|
heme binding protein 1
|
|
chrX_+_51092407
|
1.100
|
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10
|
|
chr14_+_99775185
|
1.098
|
|
YY1
|
YY1 transcription factor
|
|
chr7_-_38637513
|
1.089
|
NM_001635
NM_139316
|
AMPH
|
amphiphysin
|
|
chr9_-_23811842
|
1.087
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr17_-_74432671
|
1.085
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr12_-_105165805
|
1.083
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr15_+_61122139
|
1.074
|
|
TPM1
|
tropomyosin 1 (alpha)
|
|
chr18_-_75894902
|
1.069
|
|
|
|
|
chr18_+_58341637
|
1.068
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr6_+_3697328
|
1.068
|
|
|
|
|
chr17_-_53387493
|
1.066
|
|
CUEDC1
|
CUE domain containing 1
|
|
chr11_+_101485946
|
1.065
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr9_+_111850770
|
1.065
|
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr2_-_215382537
|
1.057
|
NM_000465
|
BARD1
|
BRCA1 associated RING domain 1
|
|
chr20_+_13924263
|
1.052
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr2_+_9532087
|
1.049
|
NM_001039613
|
IAH1
|
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae)
|
|
chr1_+_9275518
|
1.047
|
NM_025106
|
SPSB1
|
splA/ryanodine receptor domain and SOCS box containing 1
|
|
chr5_+_43228083
|
1.045
|
NM_153361
|
MGC42105
|
serine/threonine-protein kinase NIM1
|
|
chr16_-_52094613
|
1.040
|
NM_001012398
NM_022476
|
AKTIP
|
AKT interacting protein
|
|
chr2_+_233270286
|
1.040
|
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr10_-_725522
|
1.038
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr13_-_20533629
|
1.038
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr5_-_58370693
|
1.037
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_6400470
|
1.033
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr8_+_120289735
|
1.032
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr20_+_61798512
|
1.029
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr4_-_158111504
|
1.028
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr7_-_117300702
|
1.024
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chrX_+_18353623
|
1.022
|
NM_003159
|
CDKL5
|
cyclin-dependent kinase-like 5
|
|
chr1_-_20684858
|
1.022
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr22_-_49292995
|
1.019
|
NM_033200
|
LMF2
|
lipase maturation factor 2
|
|
chr6_-_3697244
|
1.011
|
NM_183373
|
C6orf145
|
chromosome 6 open reading frame 145
|
|
chr4_-_96689373
|
1.011
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr17_+_77779570
|
1.008
|
NM_001042422
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr2_+_159533329
|
1.000
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr6_-_165996031
|
0.999
|
|
LOC100132188
|
LP7097
|
|
chr22_-_41375323
|
0.997
|
NM_000398
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr22_+_49459935
|
0.990
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr8_-_101227203
|
0.989
|
NM_001029860
|
FBXO43
|
F-box protein 43
|
|
chr11_+_12355600
|
0.988
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr18_+_7557313
|
0.987
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr15_+_99237555
|
0.986
|
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr15_+_49421004
|
0.980
|
NM_181789
|
GLDN
|
gliomedin
|
|
chr11_-_11987115
|
0.978
|
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr20_-_17610704
|
0.975
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr3_-_27500810
|
0.974
|
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr11_+_118544625
|
0.969
|
NM_024618
NM_170722
|
NLRX1
|
NLR family member X1
|
|
chr9_-_21984379
|
0.968
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr17_-_15407497
|
0.968
|
NM_001135036
NM_145301
|
FAM18B2-CDRT4
FAM18B2
|
FAM18B2-CDRT4 read-through transcript
family with sequence similarity 18, member B2
|
|
chr21_-_36450448
|
0.967
|
|
LOC100506428
|
hypothetical LOC100506428
|
|
chr21_+_36993302
|
0.966
|
|
|
|
|
chr12_-_13044448
|
0.956
|
|
HEBP1
|
heme binding protein 1
|
|
chr11_-_11987198
|
0.955
|
NM_015881
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|