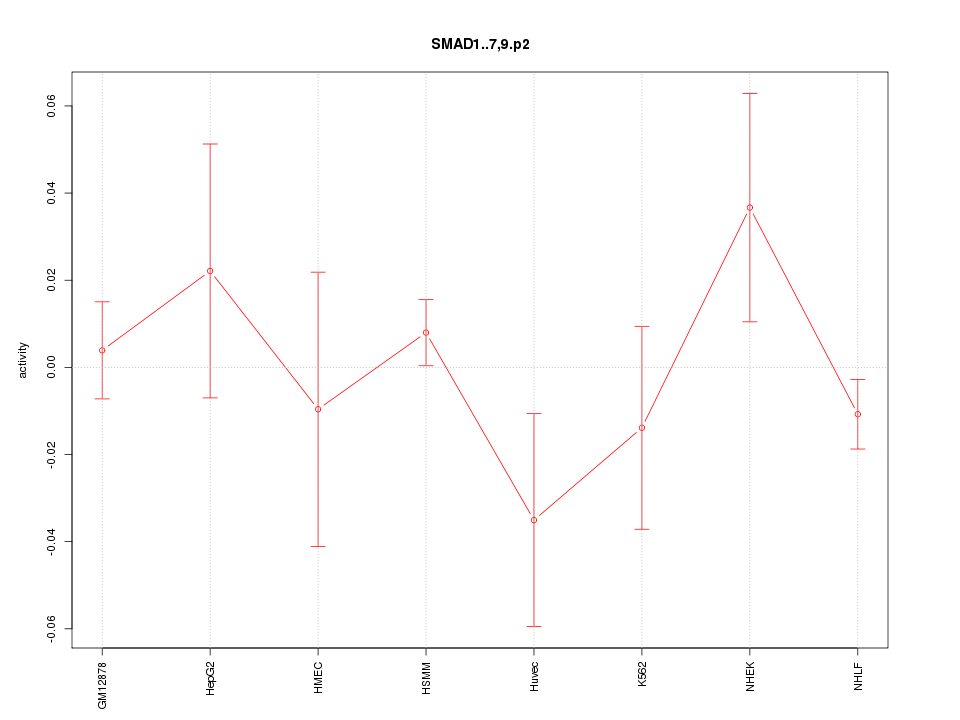
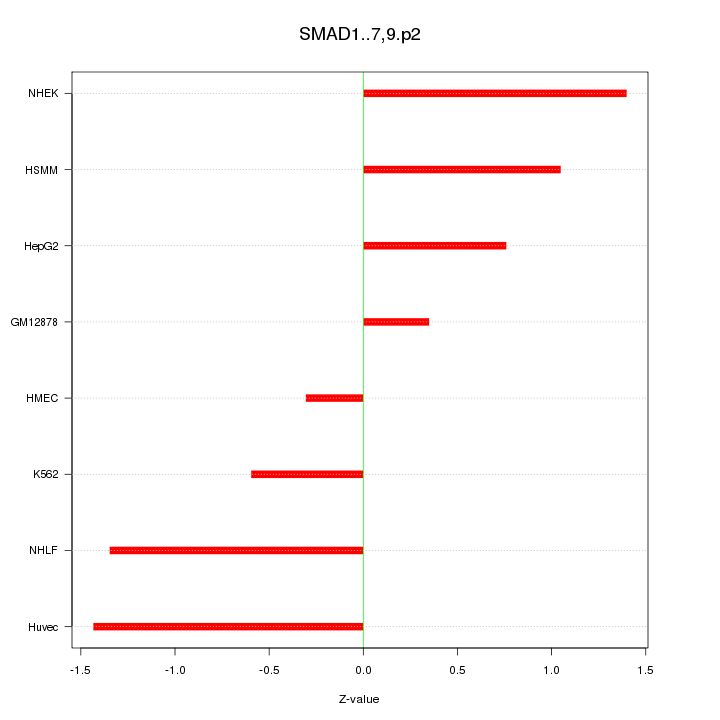
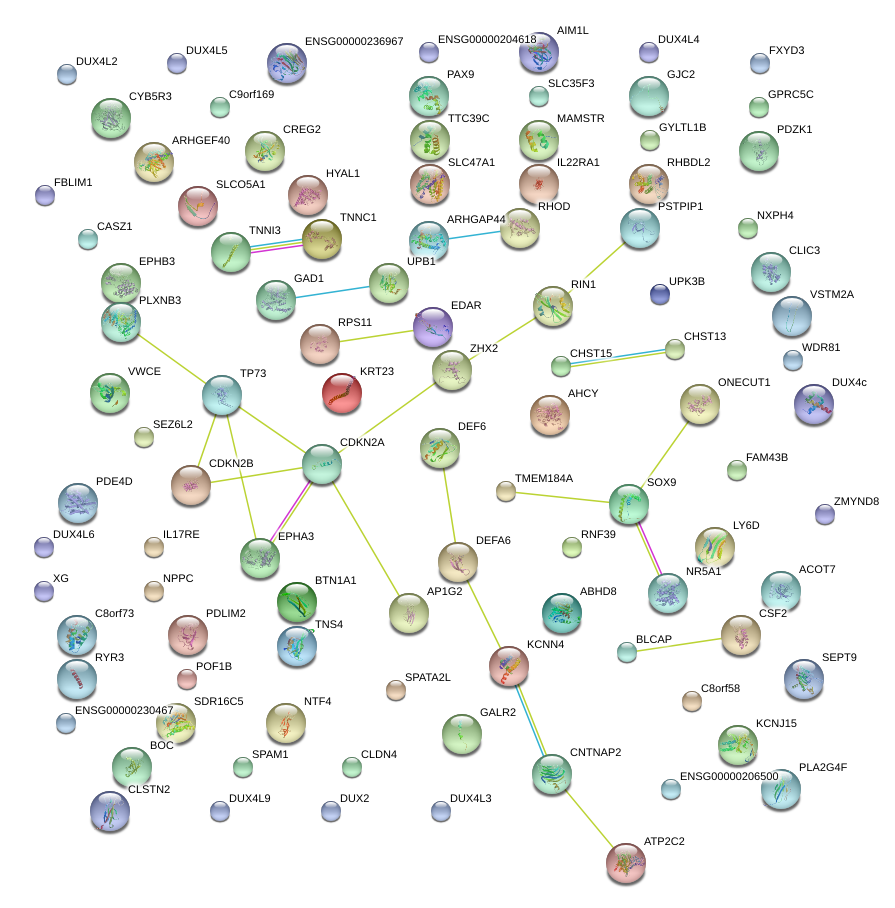
|
chr6_-_30151526
|
2.778
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr17_+_12633504
|
2.038
|
NM_014859
|
ARHGAP44
|
Rho GTPase activating protein 44
|
|
chr8_+_22493928
|
1.870
|
NM_176871
NM_198042
|
PDLIM2
|
PDZ and LIM domain 2 (mystique)
|
|
chr19_-_17275083
|
1.800
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr11_-_65860450
|
1.755
|
NM_004292
|
RIN1
|
Ras and Rab interactor 1
|
|
chr16_+_82959626
|
1.751
|
NM_014861
|
ATP2C2
|
ATPase, Ca++ transporting, type 2C, member 2
|
|
chr17_-_67628523
|
1.610
|
|
FLJ37644
|
hypothetical LOC400618
|
|
chr3_-_50324815
|
1.594
|
NM_153281
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr1_+_232416618
|
1.548
|
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chrX_+_152682772
|
1.522
|
NM_001163257
NM_005393
|
PLXNB3
|
plexin B3
|
|
chr1_+_15145001
|
1.501
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr17_+_67628749
|
1.497
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr11_+_66580884
|
1.493
|
|
RHOD
|
ras homolog gene family, member D
|
|
chr15_-_40236061
|
1.473
|
NM_213600
|
PLA2G4F
|
phospholipase A2, group IVF
|
|
chr1_-_6368324
|
1.472
|
NM_181864
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr19_+_40299058
|
1.404
|
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr3_+_9919295
|
1.377
|
NM_153483
|
IL17RE
|
interleukin 17 receptor E
|
|
chr8_-_143864933
|
1.333
|
NM_003695
|
LY6D
|
lymphocyte antigen 6 complex, locus D
|
|
chr12_+_55896844
|
1.289
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr3_-_52463070
|
1.256
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chr7_+_54577512
|
1.252
|
NM_182546
|
VSTM2A
|
V-set and transmembrane domain containing 2A
|
|
chr7_+_72883128
|
1.154
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr15_+_75074793
|
1.121
|
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr16_-_29817841
|
1.116
|
NM_001114099
NM_001114100
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chrX_-_84521320
|
1.104
|
|
POF1B
|
premature ovarian failure, 1B
|
|
chr9_-_21965131
|
1.088
|
NM_000077
NM_001195132
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr1_-_26553168
|
1.083
|
NM_001039775
|
AIM1L
|
absent in melanoma 1-like
|
|
chr17_+_69941430
|
1.074
|
NM_018653
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr14_+_20608179
|
1.053
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr1_+_15964045
|
1.052
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr1_-_24342158
|
1.050
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr22_-_41372620
|
1.043
|
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr17_+_19377732
|
1.043
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr22_+_23221114
|
1.021
|
NM_016327
|
UPB1
|
ureidopropionase, beta
|
|
chr17_+_72795567
|
1.021
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr20_-_32363268
|
1.018
|
NM_001161766
|
AHCY
|
adenosylhomocysteinase
|
|
chr2_-_232499202
|
1.007
|
NM_024409
|
NPPC
|
natriuretic peptide C
|
|
chr15_+_31390454
|
1.005
|
NM_001036
|
RYR3
|
ryanodine receptor 3
|
|
chr7_+_147667461
|
0.995
|
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr10_-_125841840
|
0.995
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr3_+_9919511
|
0.992
|
NM_001193380
NM_153480
|
IL17RE
|
interleukin 17 receptor E
|
|
chr7_+_75977680
|
0.992
|
NM_030570
NM_182684
|
UPK3B
|
uroplakin 3B
|
|
chr5_-_58370693
|
0.991
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_3558958
|
0.986
|
NM_005427
|
TP73
|
tumor protein p73
|
|
chr17_-_36346240
|
0.956
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr9_-_21964825
|
0.955
|
NM_058197
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chrX_+_2680092
|
0.944
|
NM_001141919
NM_001141920
NM_175569
|
XGPY2
XG
|
Xg pseudogene, Y-linked 2
Xg blood group
|
|
chr17_-_35911340
|
0.943
|
NM_032865
|
TNS4
|
tensin 4
|
|
chr14_+_36196523
|
0.942
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr6_+_26609427
|
0.934
|
NM_001732
|
BTN1A1
|
butyrophilin, subfamily 1, member A1
|
|
chr19_-_54258935
|
0.925
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr15_+_75074483
|
0.919
|
NM_003978
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr3_+_114414064
|
0.909
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr3_+_185762233
|
0.908
|
NM_004443
|
EPHB3
|
EPH receptor B3
|
|
chr22_-_41372920
|
0.904
|
NM_001129819
NM_001171661
NM_007326
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr1_+_144439069
|
0.902
|
NM_002614
|
PDZK1
|
PDZ domain containing 1
|
|
chr14_-_23107106
|
0.902
|
NM_003917
|
AP1G2
|
adaptor-related protein complex 1, gamma 2 subunit
|
|
chr2_+_171381317
|
0.895
|
NM_000817
NM_013445
|
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa)
|
|
chr1_-_10779291
|
0.887
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|
|
chr2_-_101370291
|
0.884
|
|
CREG2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr1_-_43523825
|
0.872
|
NM_182517
NM_001164829
|
C1orf210
|
chromosome 1 open reading frame 210
|
|
chr11_+_45900731
|
0.868
|
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr6_+_35373572
|
0.865
|
NM_022047
|
DEF6
|
differentially expressed in FDCP 6 homolog (mouse)
|
|
chr1_+_20751518
|
0.863
|
NM_207334
|
FAM43B
|
family with sequence similarity 43, member B
|
|
chr20_-_35586366
|
0.863
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr12_+_56406296
|
0.854
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr11_-_60819110
|
0.851
|
|
VWCE
|
von Willebrand factor C and EGF domains
|
|
chr19_-_48976852
|
0.849
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr9_-_126309510
|
0.841
|
NM_004959
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr15_+_75074743
|
0.840
|
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1
|
|
chr15_-_50869379
|
0.837
|
NM_004498
|
ONECUT1
|
one cut homeobox 1
|
|
chr16_-_29818334
|
0.835
|
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr3_+_127725851
|
0.834
|
NM_152889
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr20_-_45418818
|
0.820
|
NM_012408
NM_183047
NM_183048
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr2_-_108972100
|
0.817
|
NM_022336
|
EDAR
|
ectodysplasin A receptor
|
|
chr9_-_139010805
|
0.814
|
NM_004669
|
CLIC3
|
chloride intracellular channel 3
|
|
chr9_+_139239464
|
0.812
|
NM_199001
|
C9orf169
|
chromosome 9 open reading frame 169
|
|
chr3_+_141136896
|
0.809
|
|
CLSTN2
|
calsyntenin 2
|
|
chr5_+_131437383
|
0.793
|
NM_000758
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage)
|
|
chr17_+_1574782
|
0.778
|
NM_001163809
|
WDR81
|
WD repeat domain 81
|
|
chr8_-_144726064
|
0.776
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr8_-_57395207
|
0.771
|
|
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5
|
|
chr17_+_69939130
|
0.765
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr8_-_70909754
|
0.761
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr21_+_38550739
|
0.760
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr3_+_89239363
|
0.759
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr1_-_39180036
|
0.754
|
NM_017821
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila)
|
|
chr16_-_88295595
|
0.750
|
NM_152339
|
SPATA2L
|
spermatogenesis associated 2-like
|
|
chr19_-_53912025
|
0.744
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr17_+_71582486
|
0.741
|
NM_003857
|
GALR2
|
galanin receptor 2
|
|
chr10_-_102880892
|
0.738
|
NM_001085398
|
TLX1NB
|
TLX1 neighbor
|
|
chr7_-_1562272
|
0.734
|
NM_001097620
|
TMEM184A
|
transmembrane protein 184A
|
|
chr10_+_135333667
|
0.734
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr8_+_123863075
|
0.732
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr1_+_226404037
|
0.730
|
NM_020435
|
GJC2
|
gap junction protein, gamma 2, 47kDa
|
|
chr1_+_154389945
|
0.730
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr12_-_121780835
|
0.728
|
NM_032554
|
GPR81
|
G protein-coupled receptor 81
|
|
chr17_+_1574583
|
0.721
|
NM_001163811
|
WDR81
|
WD repeat domain 81
|
|
chr17_+_39741306
|
0.717
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr20_-_62052922
|
0.717
|
NM_001193379
|
UCKL1
|
uridine-cytidine kinase 1-like 1
|
|
chr1_+_27062215
|
0.711
|
NM_006142
|
SFN
|
stratifin
|
|
chr11_+_66580861
|
0.709
|
NM_014578
|
RHOD
|
ras homolog gene family, member D
|
|
chr9_-_115880281
|
0.708
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr9_+_91409746
|
0.706
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr17_-_38081967
|
0.705
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr3_+_89239494
|
0.703
|
|
EPHA3
|
EPH receptor A3
|
|
chr17_+_69939806
|
0.702
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr10_+_22765996
|
0.698
|
|
LOC100289455
|
hypothetical LOC100289455
|
|
chr12_+_106236281
|
0.696
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr8_-_134378630
|
0.694
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr2_+_171381626
|
0.693
|
|
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa)
|
|
chr20_-_57948653
|
0.693
|
NM_006242
|
PPP1R3D
|
protein phosphatase 1, regulatory (inhibitor) subunit 3D
|
|
chr8_-_10550026
|
0.692
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr1_+_26220852
|
0.691
|
NM_004455
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr10_-_104201234
|
0.689
|
NM_024886
|
C10orf95
|
chromosome 10 open reading frame 95
|
|
chr3_-_187562470
|
0.684
|
NM_001080744
NM_001080745
NM_001346
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr6_-_24466258
|
0.684
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr11_-_116692128
|
0.683
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr16_-_31054563
|
0.680
|
NM_002773
|
PRSS8
|
protease, serine, 8
|
|
chr16_-_73858179
|
0.680
|
NM_001170716
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr2_+_70981233
|
0.679
|
|
VAX2
|
ventral anterior homeobox 2
|
|
chr8_+_145714142
|
0.676
|
NM_014665
|
LRRC14
|
leucine rich repeat containing 14
|
|
chr10_+_135343649
|
0.670
|
NM_012147
|
DUX2
|
double homeobox 2
|
|
chr1_+_44171578
|
0.669
|
NM_001136215
NM_057090
NM_057091
|
ARTN
|
artemin
|
|
chr3_-_127382093
|
0.667
|
NM_012190
|
ALDH1L1
|
aldehyde dehydrogenase 1 family, member L1
|
|
chr1_+_31659246
|
0.667
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr2_-_101370355
|
0.665
|
NM_153836
|
CREG2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr6_+_118335381
|
0.655
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr17_-_40858775
|
0.655
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr5_-_79322549
|
0.652
|
NM_001010891
NM_001167741
|
MTX3
|
metaxin 3
|
|
chrX_-_132376660
|
0.652
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr22_+_36365629
|
0.649
|
NM_018957
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr1_+_26220895
|
0.645
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr19_-_60573552
|
0.644
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr2_-_219633481
|
0.639
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr20_-_62081434
|
0.635
|
NM_080621
|
SAMD10
|
sterile alpha motif domain containing 10
|
|
chr14_-_49543987
|
0.635
|
NM_001012706
|
C14orf182
|
chromosome 14 open reading frame 182
|
|
chr17_-_7238546
|
0.633
|
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr16_-_56389176
|
0.633
|
NM_001130099
|
KIFC3
|
kinesin family member C3
|
|
chr7_-_44195405
|
0.633
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chrX_+_68641802
|
0.633
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr19_-_18058632
|
0.633
|
NM_005535
NM_153701
|
IL12RB1
|
interleukin 12 receptor, beta 1
|
|
chr11_-_60475558
|
0.631
|
NM_016582
|
SLC15A3
|
solute carrier family 15, member 3
|
|
chr3_-_127382051
|
0.631
|
|
ALDH1L1
|
aldehyde dehydrogenase 1 family, member L1
|
|
chr17_-_36719030
|
0.629
|
NM_001146182
|
KRTAP16-1
|
keratin associated protein 16-1
|
|
chr10_+_135330357
|
0.626
|
NM_033178
NM_012147
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr15_+_38923470
|
0.624
|
NM_003710
NM_181642
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr2_+_70981223
|
0.623
|
NM_012476
|
VAX2
|
ventral anterior homeobox 2
|
|
chr11_-_34491878
|
0.622
|
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr2_+_242400702
|
0.620
|
NM_001167599
NM_001167602
NM_080741
|
NEU4
|
sialidase 4
|
|
chr9_+_35663852
|
0.620
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr4_+_191239247
|
0.619
|
NM_033178
NM_012147
NM_001177376
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr14_+_23527866
|
0.612
|
NM_198083
|
DHRS4
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4
dehydrogenase/reductase (SDR family) member 4 like 2
|
|
chr1_-_6341553
|
0.611
|
NM_181866
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr17_-_5078773
|
0.610
|
NM_207103
|
C17orf87
|
chromosome 17 open reading frame 87
|
|
chr4_+_191232653
|
0.608
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr19_+_49972965
|
0.607
|
NM_001130852
NM_012116
|
CBLC
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence c
|
|
chr16_-_21919928
|
0.600
|
NM_173806
|
PDZD9
|
PDZ domain containing 9
|
|
chr2_+_17585462
|
0.596
|
|
VSNL1
|
visinin-like 1
|
|
chr16_+_23221091
|
0.594
|
NM_000336
|
SCNN1B
|
sodium channel, nonvoltage-gated 1, beta
|
|
chr8_+_123863169
|
0.592
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr4_+_191229360
|
0.589
|
NM_033178
NM_012147
NM_001177376
NM_001127387
NM_001127388
NM_001127389
NM_001164467
NM_001127386
|
DUX4
DUX2
DUX4L4
DUX4L7
DUX4L6
DUX4L5
DUX4L3
DUX4L2
|
double homeobox 4
double homeobox 2
double homeobox 4 like 4
double homeobox 4 like 7
double homeobox 4 like 6
double homeobox 4 like 5
double homeobox 4 like 3
double homeobox 4 like 2
|
|
chr17_-_73867748
|
0.589
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr7_-_100331417
|
0.586
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr21_+_44818033
|
0.586
|
NM_198687
|
KRTAP10-4
|
keratin associated protein 10-4
|
|
chr22_+_29848923
|
0.586
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr9_-_115880366
|
0.586
|
|
AMBP
|
alpha-1-microglobulin/bikunin precursor
|
|
chr8_-_8276598
|
0.584
|
NM_001080826
|
SGK223
|
homolog of rat pragma of Rnd2
|
|
chr1_-_112333192
|
0.584
|
NM_004980
NM_172198
|
KCND3
|
potassium voltage-gated channel, Shal-related subfamily, member 3
|
|
chr16_+_88467518
|
0.584
|
|
TCF25
|
transcription factor 25 (basic helix-loop-helix)
|
|
chr7_+_128367961
|
0.579
|
NM_001098627
|
IRF5
|
interferon regulatory factor 5
|
|
chr3_-_48573556
|
0.578
|
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr17_+_4621924
|
0.570
|
NM_003963
|
TM4SF5
|
transmembrane 4 L six family member 5
|
|
chrX_+_21302317
|
0.569
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr8_-_145122884
|
0.566
|
NM_000445
|
PLEC
|
plectin
|
|
chr7_-_127788893
|
0.563
|
NM_001114726
NM_001174164
|
PRRT4
|
proline-rich transmembrane protein 4
|
|
chr17_+_68672705
|
0.561
|
NM_001050
|
SSTR2
|
somatostatin receptor 2
|
|
chr8_+_123863224
|
0.559
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr19_-_56164628
|
0.557
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr2_+_37425240
|
0.556
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr1_+_22224571
|
0.554
|
|
HSPC157
|
hypothetical LOC29092
|
|
chr16_-_45339605
|
0.553
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr16_-_11257539
|
0.552
|
NM_003745
|
SOCS1
|
suppressor of cytokine signaling 1
|
|
chr12_+_6292077
|
0.550
|
NM_001144857
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr9_-_99994704
|
0.548
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr1_+_178466064
|
0.548
|
NM_033343
|
LHX4
|
LIM homeobox 4
|
|
chr19_-_48977247
|
0.546
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr1_-_181826633
|
0.545
|
NM_001127651
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr19_+_702108
|
0.542
|
NM_173481
|
C19orf21
|
chromosome 19 open reading frame 21
|
|
chr15_+_38923914
|
0.542
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr2_-_108972048
|
0.539
|
|
EDAR
|
ectodysplasin A receptor
|
|
chr22_+_31527678
|
0.539
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr4_-_78038025
|
0.539
|
NM_001029870
|
ANKRD56
|
ankyrin repeat domain 56
|
|
chr19_+_45807909
|
0.537
|
|
|
|
|
chr16_+_22732982
|
0.530
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chrX_-_74062005
|
0.528
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr6_+_19946591
|
0.527
|
|
|
|
|
chr15_-_61680003
|
0.526
|
|
LOC100130855
|
hypothetical LOC100130855
|
|
chr14_-_74491878
|
0.525
|
NM_002632
|
PGF
|
placental growth factor
|
|
chr20_-_34835546
|
0.524
|
NM_001145316
NM_024918
NM_001145315
NM_001145317
NM_001145318
|
DSN1
|
DSN1, MIND kinetochore complex component, homolog (S. cerevisiae)
|