|
chr6_-_72187168
|
2.458
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr13_-_98428244
|
1.563
|
NM_001130048
NM_001130050
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr8_-_120033240
|
1.555
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr7_-_27171673
|
1.513
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr12_-_6535481
|
1.490
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr7_-_27171648
|
1.487
|
|
HOXA9
|
homeobox A9
|
|
chr7_-_27171600
|
1.381
|
|
HOXA9
|
homeobox A9
|
|
chr12_-_6103945
|
1.236
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chrX_-_83644054
|
1.235
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr3_+_100840129
|
1.220
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr1_-_68470585
|
1.148
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr7_+_27102197
|
1.136
|
|
|
|
|
chr14_+_85066265
|
1.135
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr21_-_27261276
|
1.135
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr4_+_156899575
|
1.092
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr14_+_85066207
|
1.088
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr19_-_45888259
|
1.052
|
NM_004756
|
NUMBL
|
numb homolog (Drosophila)-like
|
|
chr2_-_56266408
|
1.049
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr7_-_27102049
|
0.996
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr7_+_134114957
|
0.987
|
|
CALD1
|
caldesmon 1
|
|
chr8_+_55533047
|
0.986
|
NM_022454
|
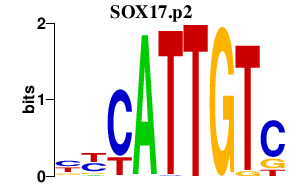
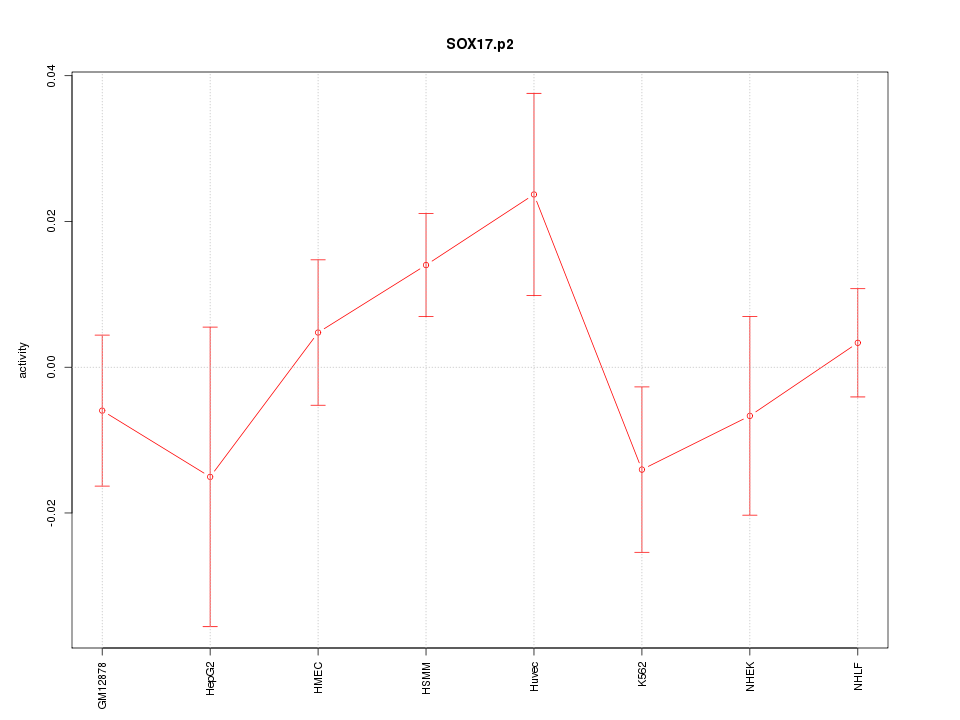
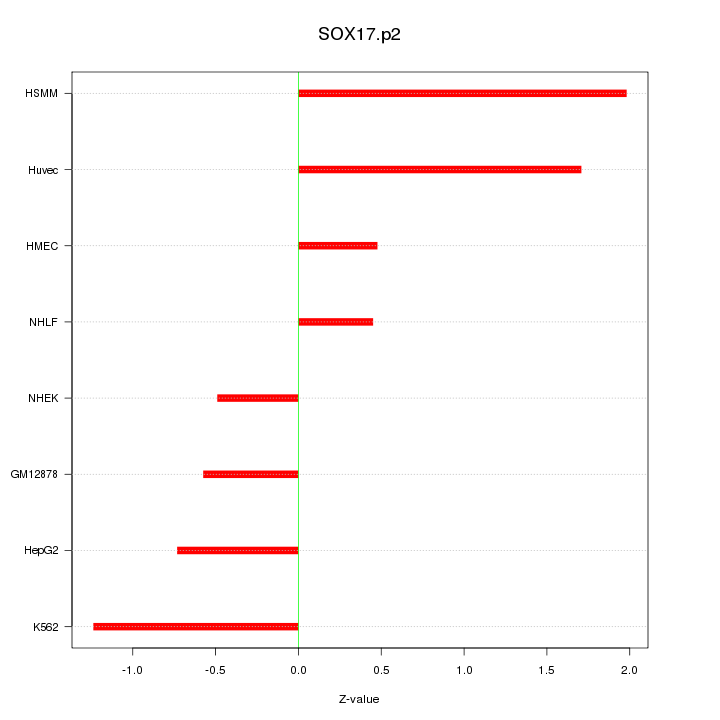
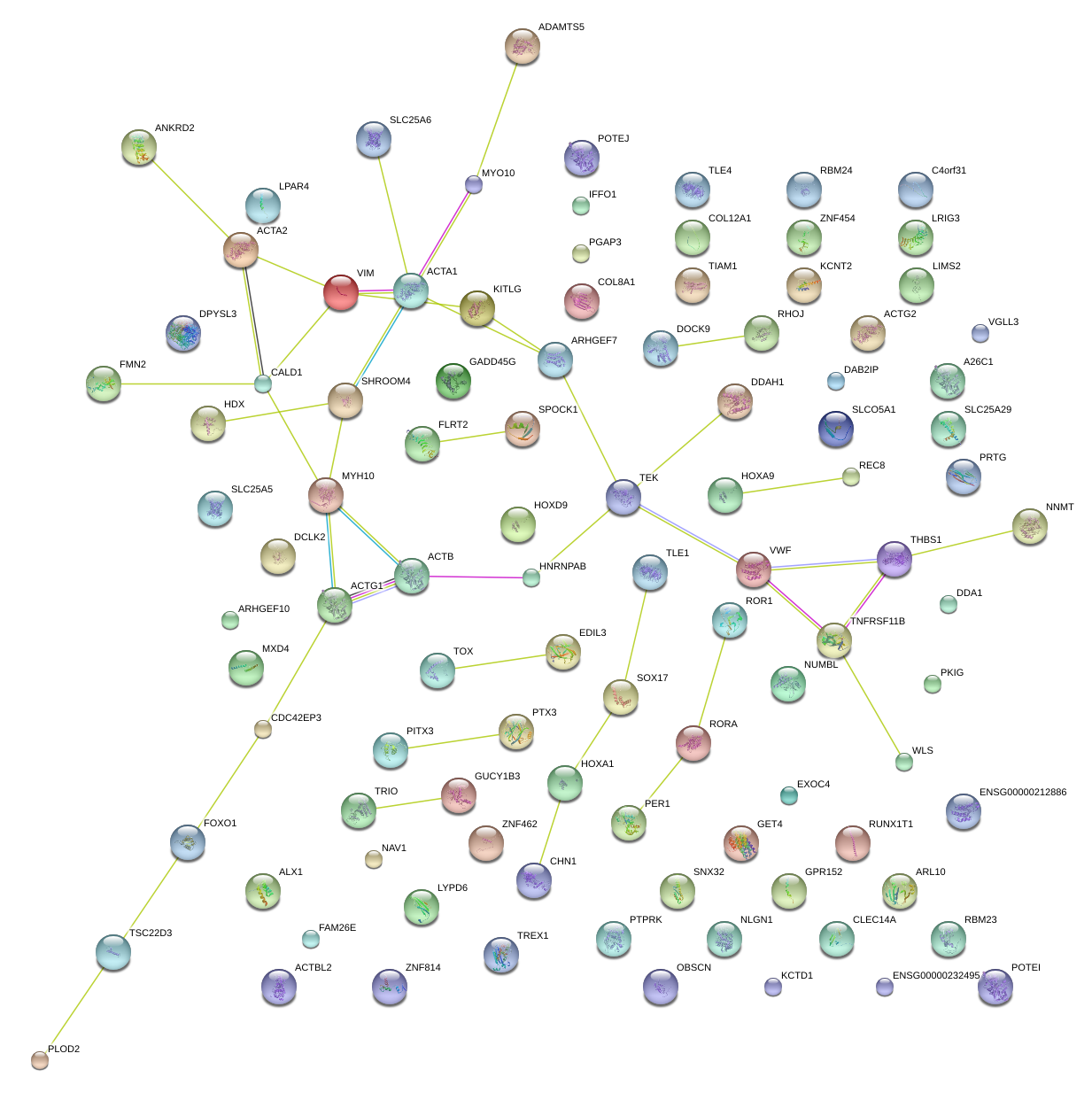
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr8_-_93184629
|
0.977
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr15_-_53822468
|
0.962
|
NM_173814
|
PRTG
|
protogenin
|
|
chr17_-_7996409
|
0.957
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr7_+_148613304
|
0.954
|
|
LOC155060
|
AI894139 pseudogene
|
|
chr11_+_113672378
|
0.926
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr7_+_27102508
|
0.925
|
|
|
|
|
chrX_+_77889861
|
0.885
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr3_-_87122958
|
0.880
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr6_-_75972473
|
0.878
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr3_-_87122936
|
0.877
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr11_+_113672407
|
0.872
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr14_-_99842519
|
0.866
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chrX_-_50403412
|
0.864
|
|
SHROOM4
|
shroom family member 4
|
|
chr5_-_146813343
|
0.861
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr14_-_37795001
|
0.853
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr21_-_31853160
|
0.851
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr4_+_151218862
|
0.849
|
NM_001040260
NM_001040261
|
DCLK2
|
doublecortin-like kinase 2
|
|
chr3_+_158637273
|
0.848
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr1_+_64012154
|
0.843
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr2_-_37752272
|
0.843
|
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr4_-_2233606
|
0.830
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr8_+_1759502
|
0.828
|
NM_014629
|
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr3_+_48482625
|
0.821
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr12_-_57600336
|
0.808
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr7_+_134114912
|
0.806
|
|
CALD1
|
caldesmon 1
|
|
chr12_+_84198015
|
0.798
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr4_-_122213018
|
0.796
|
NM_024574
|
C4orf31
|
chromosome 4 open reading frame 31
|
|
chr17_-_8474733
|
0.791
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr5_-_83716346
|
0.789
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr2_+_149895274
|
0.786
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr5_-_16989290
|
0.782
|
|
MYO10
|
myosin X
|
|
chr8_-_60194099
|
0.780
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr12_-_57600564
|
0.778
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr5_+_175725050
|
0.778
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr18_-_22382497
|
0.778
|
NM_001142730
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr14_-_37795290
|
0.769
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr13_+_110653407
|
0.766
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr5_+_178300799
|
0.763
|
NM_001178089
NM_001178090
NM_182594
|
ZNF454
|
zinc finger protein 454
|
|
chr1_-_194844299
|
0.760
|
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr10_+_22765996
|
0.759
|
|
LOC100289455
|
hypothetical LOC100289455
|
|
chr1_+_238321794
|
0.753
|
NM_020066
|
FMN2
|
formin 2
|
|
chr11_+_65357972
|
0.748
|
NM_152760
|
SNX32
|
sorting nexin 32
|
|
chr9_+_108665168
|
0.743
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr5_-_16989371
|
0.740
|
NM_012334
|
MYO10
|
myosin X
|
|
chr20_+_42644633
|
0.736
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr1_-_197173159
|
0.730
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr5_-_136862317
|
0.722
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr7_-_5535726
|
0.722
|
|
ACTB
|
actin, beta
|
|
chr8_-_70908130
|
0.720
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr3_+_174598937
|
0.713
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr5_+_14493905
|
0.704
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr5_-_83715947
|
0.696
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr1_+_199975674
|
0.694
|
|
NAV1
|
neuron navigator 1
|
|
chr14_+_23711044
|
0.693
|
NM_001048205
NM_005132
|
REC8
|
REC8 homolog (yeast)
|
|
chr5_-_136862130
|
0.691
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr9_+_123501186
|
0.686
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr6_-_128883125
|
0.673
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr6_+_17390910
|
0.672
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr19_-_63092099
|
0.667
|
NM_001144989
|
ZNF814
|
zinc finger protein 814
|
|
chr7_+_882766
|
0.659
|
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr7_+_882704
|
0.657
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr13_-_40138607
|
0.651
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr18_-_29412148
|
0.643
|
|
|
|
|
chr14_+_62740878
|
0.638
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr3_-_147361912
|
0.635
|
NM_000935
NM_182943
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr2_-_175578156
|
0.633
|
NM_001025201
NM_001822
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr14_-_22458134
|
0.633
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr9_+_91409746
|
0.628
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr2_+_149894744
|
0.618
|
NM_001195685
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chrX_-_1470862
|
0.617
|
NM_001636
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6
|
|
chr19_+_17281352
|
0.616
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr9_+_81377419
|
0.615
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr12_-_87498225
|
0.613
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr14_-_22458220
|
0.610
|
NM_001077351
NM_001077352
NM_018107
|
RBM23
|
RNA binding motif protein 23
|
|
chr10_+_17312654
|
0.603
|
|
VIM
|
vimentin
|
|
chr2_+_12775814
|
0.602
|
|
|
|
|
chr1_-_85703198
|
0.599
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr6_+_116939500
|
0.594
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr7_+_134114701
|
0.594
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr11_-_66976775
|
0.593
|
NM_206997
|
GPR152
|
G protein-coupled receptor 152
|
|
chr5_+_177564139
|
0.592
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr15_+_37660568
|
0.591
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr9_+_27099138
|
0.590
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr1_-_194844061
|
0.588
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr2_-_128138435
|
0.587
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chrX_-_106846253
|
0.576
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr14_-_22458199
|
0.574
|
|
RBM23
|
RNA binding motif protein 23
|
|
chr19_+_54723044
|
0.572
|
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr4_-_138673014
|
0.570
|
|
PCDH18
|
protocadherin 18
|
|
chr11_+_17697685
|
0.568
|
NM_002478
|
MYOD1
|
myogenic differentiation 1
|
|
chrX_+_15428820
|
0.567
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr10_+_124211353
|
0.566
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr4_-_2233504
|
0.566
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr15_-_42274760
|
0.563
|
|
FRMD5
|
FERM domain containing 5
|
|
chr8_-_101187346
|
0.562
|
|
RGS22
|
regulator of G-protein signaling 22
|
|
chr13_-_32658215
|
0.561
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_148593911
|
0.561
|
|
|
|
|
chr2_-_45090025
|
0.560
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr19_+_47593137
|
0.560
|
|
|
|
|
chr4_+_119174947
|
0.559
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr3_+_106568334
|
0.557
|
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr3_-_107070400
|
0.555
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr12_-_24606616
|
0.551
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr5_+_40715799
|
0.550
|
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr19_+_63482118
|
0.550
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chr19_+_17281216
|
0.548
|
NM_024050
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr10_+_124210980
|
0.547
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr3_+_48482213
|
0.541
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr14_+_76298693
|
0.540
|
|
VASH1
|
vasohibin 1
|
|
chr15_-_32875040
|
0.539
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr5_+_177564102
|
0.536
|
NM_004499
NM_031266
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr21_+_33320080
|
0.535
|
NM_005806
|
OLIG2
|
oligodendrocyte lineage transcription factor 2
|
|
chr3_-_11736946
|
0.534
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr3_+_106568243
|
0.528
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr9_+_81377289
|
0.528
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr5_+_40715778
|
0.525
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr19_+_17281374
|
0.525
|
|
DDA1
|
DET1 and DDB1 associated 1
|
|
chr5_-_111782637
|
0.524
|
NM_022140
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A
|
|
chr11_+_13646727
|
0.523
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr11_+_128068982
|
0.521
|
NM_002017
|
FLI1
|
Friend leukemia virus integration 1
|
|
chr14_+_23710924
|
0.520
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr16_+_580092
|
0.519
|
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr5_-_124108672
|
0.518
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr15_-_43457916
|
0.518
|
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr10_-_116434102
|
0.517
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr8_-_6407882
|
0.511
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr3_+_106568430
|
0.510
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr4_+_156900053
|
0.510
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr7_+_68702516
|
0.507
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr19_-_58449919
|
0.506
|
NM_182609
|
ZNF677
|
zinc finger protein 677
|
|
chr7_-_117300702
|
0.506
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr8_+_60194328
|
0.500
|
|
|
|
|
chr1_+_175406685
|
0.498
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr10_+_3099708
|
0.498
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr6_-_134680888
|
0.497
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr17_-_1478293
|
0.495
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr1_-_152431175
|
0.493
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr1_+_43539239
|
0.492
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr6_-_56824371
|
0.489
|
|
DST
|
dystonin
|
|
chr16_+_580057
|
0.488
|
NM_001172665
NM_001172666
|
RAB40C
|
RAB40C, member RAS oncogene family
|
|
chr5_+_53787187
|
0.485
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr7_-_5535432
|
0.485
|
|
ACTB
|
actin, beta
|
|
chr8_+_69027143
|
0.484
|
NM_024870
NM_025170
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr15_-_70277163
|
0.482
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr5_+_177564140
|
0.480
|
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B
|
|
chr1_+_153560841
|
0.480
|
NM_014328
|
RUSC1
|
RUN and SH3 domain containing 1
|
|
chr5_+_139761579
|
0.480
|
NM_020690
NM_001197030
NM_017747
NM_017978
NM_024668
|
ANKHD1-EIF4EBP3
ANKHD1
|
ANKHD1-EIF4EBP3 readthrough
ankyrin repeat and KH domain containing 1
|
|
chr10_-_125843092
|
0.480
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr1_+_45565126
|
0.479
|
NM_032756
|
HPDL
|
4-hydroxyphenylpyruvate dioxygenase-like
|
|
chr12_+_77782648
|
0.478
|
|
SYT1
|
synaptotagmin I
|
|
chr10_-_116434364
|
0.478
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr6_-_85529884
|
0.476
|
|
TBX18
|
T-box 18
|
|
chr17_-_59817456
|
0.472
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr13_+_94052022
|
0.470
|
NM_180989
|
GPR180
|
G protein-coupled receptor 180
|
|
chr7_-_13995811
|
0.469
|
|
ETV1
|
ets variant 1
|
|
chr12_-_24946495
|
0.467
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr10_+_123912930
|
0.467
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr3_+_44665220
|
0.464
|
NM_003420
|
ZNF35
|
zinc finger protein 35
|
|
chr8_-_70907957
|
0.463
|
NM_001146009
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr8_-_38444398
|
0.463
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr4_-_138673078
|
0.462
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr10_+_104667991
|
0.461
|
NM_017649
NM_199076
NM_199077
|
CNNM2
|
cyclin M2
|
|
chr11_+_13646806
|
0.459
|
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr1_+_98899734
|
0.457
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chrX_+_136475979
|
0.457
|
NM_003413
|
ZIC3
|
Zic family member 3 (odd-paired homolog, Drosophila)
|
|
chr6_+_142665057
|
0.457
|
|
GPR126
|
G protein-coupled receptor 126
|
|
chr13_-_29067795
|
0.456
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr2_-_71307705
|
0.456
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr11_-_4585947
|
0.454
|
NM_018073
|
TRIM68
|
tripartite motif containing 68
|
|
chr6_+_17390273
|
0.453
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr7_-_27186400
|
0.453
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr2_-_144991386
|
0.450
|
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr11_-_74914514
|
0.450
|
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr3_-_189354510
|
0.450
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr7_-_130891861
|
0.448
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr14_+_102870301
|
0.446
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr17_-_59817575
|
0.446
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr11_-_27679617
|
0.445
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr5_+_140835727
|
0.444
|
NM_002588
NM_032402
NM_032403
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|