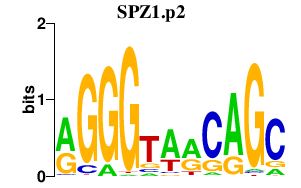
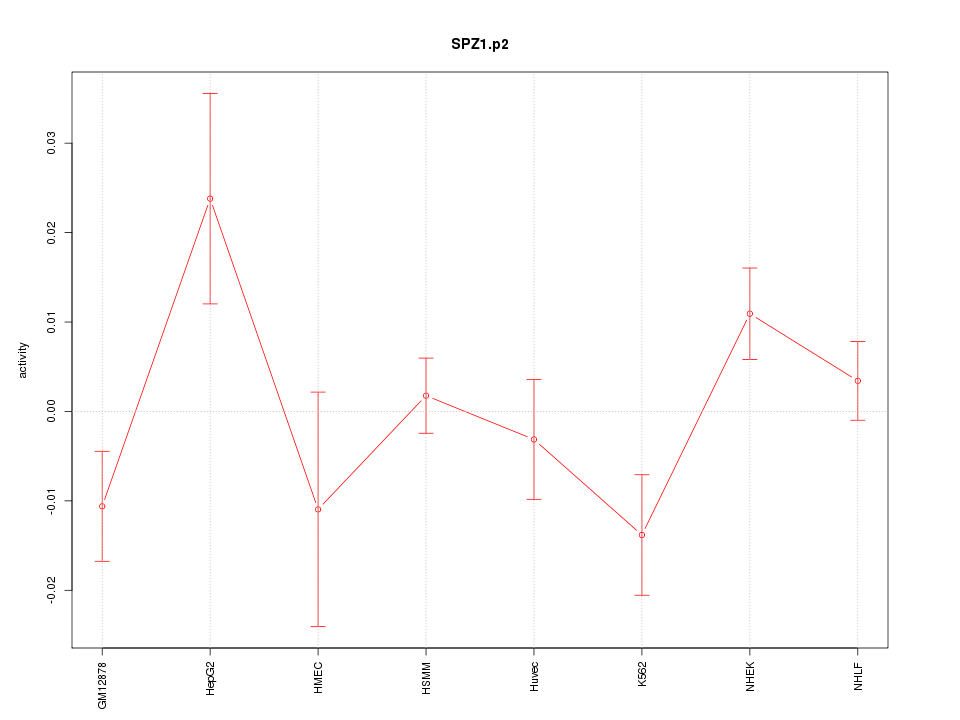
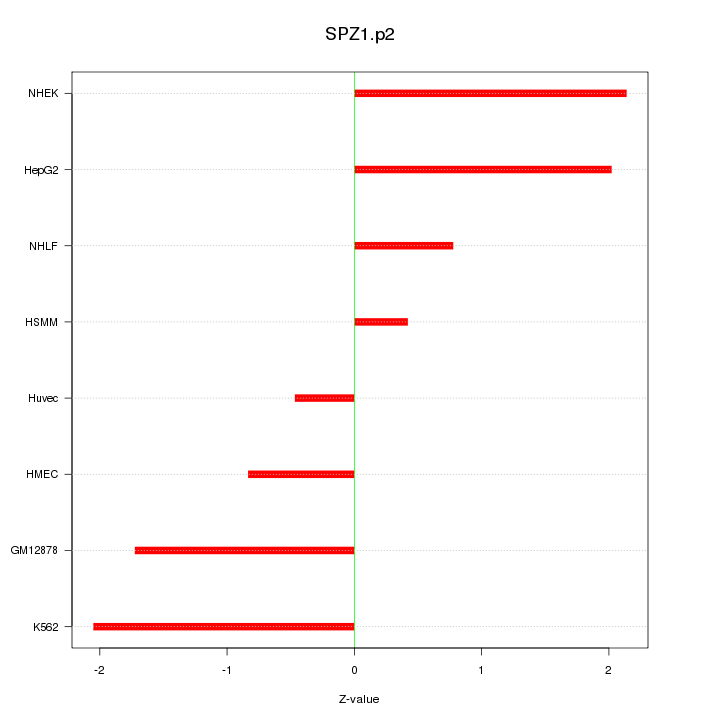
|
chr16_+_29819647
|
1.759
|
NM_181718
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1
|
|
chr16_+_29373365
|
1.660
|
NM_001014999
NM_001015000
NM_024044
NM_178044
|
SLX1A-SULT1A3
SLX1A
SLX1B
|
SLX1A-SULT1A3 read-through transcript
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae)
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae)
|
|
chr14_+_23937766
|
1.560
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr10_+_123862430
|
1.506
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chrX_+_105856531
|
1.468
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr20_-_30535045
|
1.460
|
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr12_+_64504835
|
1.393
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr16_-_66535457
|
1.387
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr20_-_22513050
|
1.361
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr5_+_129268352
|
1.349
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr22_-_19122047
|
1.294
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr14_-_24588916
|
1.259
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr3_-_150858274
|
1.214
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr2_-_42873179
|
1.205
|
NM_012205
|
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr4_-_118226119
|
1.157
|
NM_152402
|
TRAM1L1
|
translocation associated membrane protein 1-like 1
|
|
chr18_-_50777634
|
1.148
|
NM_025214
|
CCDC68
|
coiled-coil domain containing 68
|
|
chr9_+_115957537
|
1.148
|
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr12_-_47468985
|
1.132
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr1_-_150070849
|
1.114
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr17_+_72795567
|
1.112
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr1_+_199130571
|
1.090
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr2_+_70981223
|
1.086
|
NM_012476
|
VAX2
|
ventral anterior homeobox 2
|
|
chr7_+_94374862
|
1.076
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr7_+_45894480
|
1.061
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr2_-_220144254
|
1.033
|
|
OBSL1
|
obscurin-like 1
|
|
chr17_-_37144408
|
1.025
|
NM_001079870
NM_001079871
NM_177977
|
HAP1
|
huntingtin-associated protein 1
|
|
chr9_-_85342868
|
1.008
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr17_-_77824849
|
1.001
|
NM_001893
NM_139062
|
CSNK1D
|
casein kinase 1, delta
|
|
chr17_+_71582486
|
0.992
|
NM_003857
|
GALR2
|
galanin receptor 2
|
|
chr6_-_119712596
|
0.980
|
NM_005907
|
MAN1A1
|
mannosidase, alpha, class 1A, member 1
|
|
chr2_+_70981233
|
0.978
|
|
VAX2
|
ventral anterior homeobox 2
|
|
chr4_-_158111995
|
0.960
|
NM_016205
|
PDGFC
|
platelet derived growth factor C
|
|
chr9_-_106730157
|
0.935
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr2_+_54536780
|
0.930
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr6_-_128883125
|
0.926
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr3_+_125786195
|
0.925
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr11_-_2116775
|
0.922
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr6_-_116488458
|
0.916
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr2_-_219633481
|
0.906
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr1_-_159459999
|
0.904
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr3_+_152287365
|
0.898
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr9_+_135389698
|
0.893
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr11_-_116168316
|
0.892
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr2_+_119905812
|
0.887
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr1_-_20684858
|
0.878
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr5_+_80292313
|
0.877
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr22_+_29122932
|
0.858
|
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr1_+_15955719
|
0.850
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr16_-_51138306
|
0.848
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr11_-_70641267
|
0.838
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr4_+_184257337
|
0.825
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr5_+_80292213
|
0.824
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chrX_-_132947141
|
0.822
|
|
GPC3
|
glypican 3
|
|
chr14_-_22974707
|
0.813
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr12_+_106236281
|
0.808
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chrX_-_132947290
|
0.801
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr20_-_17610704
|
0.799
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr1_-_24307514
|
0.798
|
|
MYOM3
|
myomesin family, member 3
|
|
chr1_+_151917674
|
0.797
|
NM_000906
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A)
|
|
chr3_-_187562470
|
0.792
|
NM_001080744
NM_001080745
NM_001346
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr9_-_4289574
|
0.785
|
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr16_-_66535923
|
0.784
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr20_-_22514092
|
0.782
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr12_+_50686990
|
0.782
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr14_-_23686269
|
0.782
|
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta)
|
|
chr10_-_101680645
|
0.778
|
|
DNMBP
|
dynamin binding protein
|
|
chr6_+_43036442
|
0.771
|
NM_018960
|
GNMT
|
glycine N-methyltransferase
|
|
chr12_-_48017223
|
0.768
|
NM_001008223
|
C1QL4
|
complement component 1, q subcomponent-like 4
|
|
chr2_+_242400702
|
0.763
|
NM_001167599
NM_001167602
NM_080741
|
NEU4
|
sialidase 4
|
|
chr15_-_68933481
|
0.761
|
NM_018357
NM_197958
|
LARP6
|
La ribonucleoprotein domain family, member 6
|
|
chr17_+_33761234
|
0.752
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr20_+_3399615
|
0.752
|
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr20_-_22512893
|
0.751
|
|
FOXA2
|
forkhead box A2
|
|
chr4_-_187884721
|
0.751
|
|
|
|
|
chr2_-_216944911
|
0.744
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr2_-_192767844
|
0.744
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr7_+_113513600
|
0.734
|
|
FOXP2
|
forkhead box P2
|
|
chr4_-_158111504
|
0.734
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr4_-_158111916
|
0.730
|
|
PDGFC
|
platelet derived growth factor C
|
|
chr19_+_39664706
|
0.724
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr14_-_22596522
|
0.717
|
NM_022478
NM_144985
|
CDH24
|
cadherin 24, type 2
|
|
chr10_-_105411339
|
0.716
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr4_-_22126437
|
0.711
|
NM_145290
|
GPR125
|
G protein-coupled receptor 125
|
|
chr17_+_35425139
|
0.708
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chr2_+_171280944
|
0.704
|
|
SP5
|
Sp5 transcription factor
|
|
chr12_+_56406296
|
0.698
|
|
LOC100130776
|
hypothetical LOC100130776
|
|
chr16_-_4778317
|
0.696
|
NM_001154458
NM_144605
|
SEPT12
|
septin 12
|
|
chr16_-_49742008
|
0.692
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr2_-_36384
|
0.688
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr3_+_106568430
|
0.687
|
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr20_-_42457717
|
0.684
|
|
|
|
|
chr14_+_36196523
|
0.680
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr7_+_29200850
|
0.680
|
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr6_-_30151526
|
0.679
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr9_+_70926009
|
0.674
|
NM_001170414
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr12_-_6535481
|
0.673
|
NM_001039670
NM_001193457
NM_080730
|
IFFO1
|
intermediate filament family orphan 1
|
|
chr17_+_45405128
|
0.673
|
NM_001934
|
DLX4
|
distal-less homeobox 4
|
|
chr14_+_68796621
|
0.670
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr9_+_115958051
|
0.669
|
NM_032888
|
COL27A1
|
collagen, type XXVII, alpha 1
|
|
chr11_-_124272891
|
0.667
|
NM_019055
|
ROBO4
|
roundabout homolog 4, magic roundabout (Drosophila)
|
|
chr12_+_7174230
|
0.663
|
NM_014718
|
CLSTN3
|
calsyntenin 3
|
|
chr16_-_49742651
|
0.661
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr3_+_112273352
|
0.660
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr16_+_56211410
|
0.654
|
NM_001145770
NM_005682
|
GPR56
|
G protein-coupled receptor 56
|
|
chr17_-_33179118
|
0.649
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr14_+_89597860
|
0.644
|
NM_022054
|
KCNK13
|
potassium channel, subfamily K, member 13
|
|
chr10_-_105604942
|
0.643
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr6_-_138231006
|
0.638
|
|
|
|
|
chr5_+_115326035
|
0.633
|
NM_173800
|
AQPEP
|
laeverin
|
|
chrX_-_152889815
|
0.624
|
NM_005334
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr4_-_1156362
|
0.622
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr2_-_20075929
|
0.621
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr4_-_15694407
|
0.620
|
|
PROM1
|
prominin 1
|
|
chr2_-_20075889
|
0.620
|
|
MATN3
|
matrilin 3
|
|
chr18_-_22383066
|
0.619
|
NM_001136205
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr14_-_76677711
|
0.616
|
|
ZDHHC22
|
zinc finger, DHHC-type containing 22
|
|
chr17_+_41217435
|
0.616
|
|
CRHR1
|
corticotropin releasing hormone receptor 1
|
|
chr13_+_92677012
|
0.615
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr1_-_151848039
|
0.609
|
|
S100A16
|
S100 calcium binding protein A16
|
|
chr7_+_78920846
|
0.609
|
|
LOC100505881
|
hypothetical LOC100505881
|
|
chr17_-_33178987
|
0.608
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr12_-_78608154
|
0.607
|
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr20_-_9767406
|
0.607
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr6_+_96570565
|
0.606
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr3_+_214668
|
0.605
|
|
|
|
|
chr19_+_52215982
|
0.604
|
NM_002517
|
NPAS1
|
neuronal PAS domain protein 1
|
|
chr15_-_73792234
|
0.601
|
NM_001897
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr14_-_22516271
|
0.601
|
NM_198086
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr12_-_49878216
|
0.601
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr2_-_160972606
|
0.601
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr5_+_79021401
|
0.599
|
NM_153610
|
CMYA5
|
cardiomyopathy associated 5
|
|
chr17_-_36469869
|
0.598
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr2_-_165186603
|
0.597
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr11_+_2116486
|
0.595
|
|
|
|
|
chr7_-_44195405
|
0.593
|
NM_000162
|
GCK
|
glucokinase (hexokinase 4)
|
|
chr3_-_184316553
|
0.590
|
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha)
|
|
chr6_-_24466258
|
0.587
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr22_-_22970850
|
0.587
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr14_-_59407103
|
0.577
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr6_+_32090516
|
0.577
|
NM_007293
NM_001002029
|
C4A
C4B
|
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr12_+_48026955
|
0.577
|
|
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22
|
|
chr13_-_33148899
|
0.574
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr16_-_14695930
|
0.573
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chrX_-_132376660
|
0.573
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr21_-_36774257
|
0.573
|
NM_001146079
NM_144492
|
CLDN14
|
claudin 14
|
|
chr2_+_223625105
|
0.572
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr2_+_46378405
|
0.567
|
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr12_-_24606616
|
0.566
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr17_-_40858775
|
0.560
|
NM_199282
|
ARHGAP27
|
Rho GTPase activating protein 27
|
|
chr9_-_106730256
|
0.558
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr16_+_65765256
|
0.556
|
NM_003946
NM_001185057
|
NOL3
|
nucleolar protein 3 (apoptosis repressor with CARD domain)
|
|
chr14_+_23527866
|
0.555
|
NM_198083
|
DHRS4
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4
dehydrogenase/reductase (SDR family) member 4 like 2
|
|
chr9_-_35105844
|
0.554
|
NM_025182
|
KIAA1539
|
KIAA1539
|
|
chr20_+_31334601
|
0.553
|
NM_033197
|
C20orf114
|
chromosome 20 open reading frame 114
|
|
chr6_-_31954730
|
0.552
|
|
SLC44A4
|
solute carrier family 44, member 4
|
|
chr17_+_4434024
|
0.550
|
NM_198501
|
SMTNL2
|
smoothelin-like 2
|
|
chr17_+_39603599
|
0.550
|
NM_080863
|
ASB16
|
ankyrin repeat and SOCS box containing 16
|
|
chr7_+_29200636
|
0.550
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr11_+_101488380
|
0.544
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr15_+_77511912
|
0.542
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr15_-_72804763
|
0.541
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr17_-_35017650
|
0.540
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr10_-_71753684
|
0.538
|
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr7_-_103417198
|
0.537
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr8_+_30361485
|
0.534
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr5_+_5193442
|
0.534
|
NM_139056
|
ADAMTS16
|
ADAM metallopeptidase with thrombospondin type 1 motif, 16
|
|
chr17_-_74231266
|
0.533
|
|
CYTH1
|
cytohesin 1
|
|
chr15_+_58083798
|
0.531
|
|
FOXB1
|
forkhead box B1
|
|
chr16_+_56211150
|
0.530
|
|
GPR56
|
G protein-coupled receptor 56
|
|
chr22_+_39404940
|
0.528
|
NM_005297
|
MCHR1
|
melanin-concentrating hormone receptor 1
|
|
chr9_-_106705867
|
0.524
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr15_-_73792150
|
0.523
|
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr2_+_109729176
|
0.522
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr17_-_67628523
|
0.522
|
|
FLJ37644
|
hypothetical LOC400618
|
|
chr9_-_35553895
|
0.521
|
NM_001099951
NM_001164310
|
FAM166B
|
family with sequence similarity 166, member B
|
|
chr19_+_50373842
|
0.520
|
NM_212550
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3
|
|
chr2_-_165186232
|
0.519
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr6_+_43151922
|
0.518
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr16_+_30667120
|
0.515
|
NM_000294
NM_001172432
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis)
|
|
chr8_-_144726064
|
0.515
|
NM_001100878
|
C8orf73
|
chromosome 8 open reading frame 73
|
|
chr7_-_99411573
|
0.515
|
NM_001185
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding
|
|
chr7_+_100792841
|
0.515
|
NM_133457
|
EMID2
|
EMI domain containing 2
|
|
chr2_-_127580975
|
0.514
|
NM_004305
NM_139343
NM_139344
NM_139345
NM_139346
NM_139347
NM_139348
NM_139349
NM_139350
NM_139351
|
BIN1
|
bridging integrator 1
|
|
chr15_-_71448194
|
0.512
|
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr14_-_22891263
|
0.508
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr11_-_35503720
|
0.508
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr1_-_21868380
|
0.507
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr2_-_174538197
|
0.507
|
|
SP3
|
Sp3 transcription factor
|
|
chr14_-_26136799
|
0.507
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr8_-_145613034
|
0.507
|
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr12_-_6680053
|
0.503
|
NM_153685
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
chr11_-_124127295
|
0.501
|
NM_014312
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr10_-_118419459
|
0.500
|
NM_144661
|
C10orf82
|
chromosome 10 open reading frame 82
|
|
chr8_-_125809831
|
0.499
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr19_+_49972965
|
0.499
|
NM_001130852
NM_012116
|
CBLC
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence c
|
|
chr6_+_17390273
|
0.498
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr16_-_52877868
|
0.497
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr2_-_86889029
|
0.497
|
NM_001145873
|
CD8A
|
CD8a molecule
|
|
chr17_-_74432671
|
0.497
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr2_-_192767494
|
0.495
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|