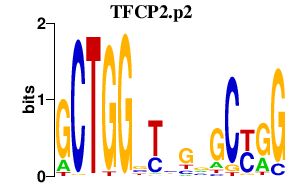
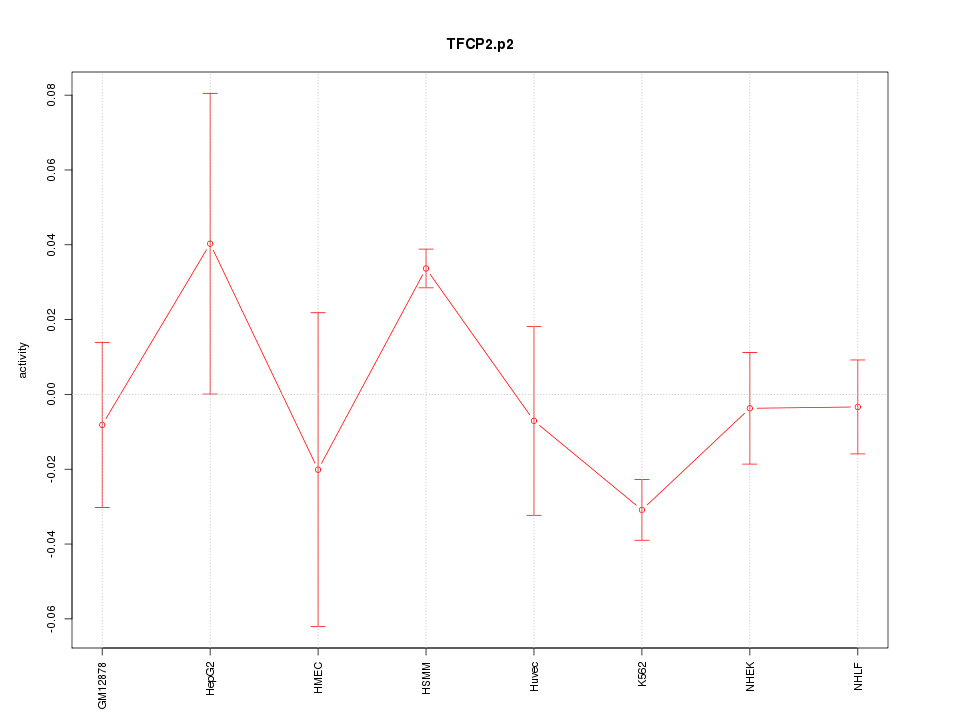
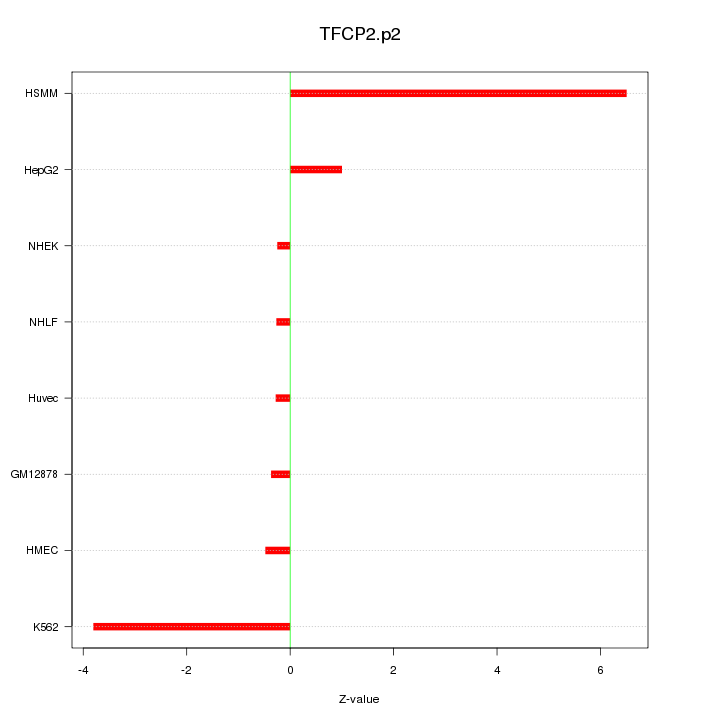
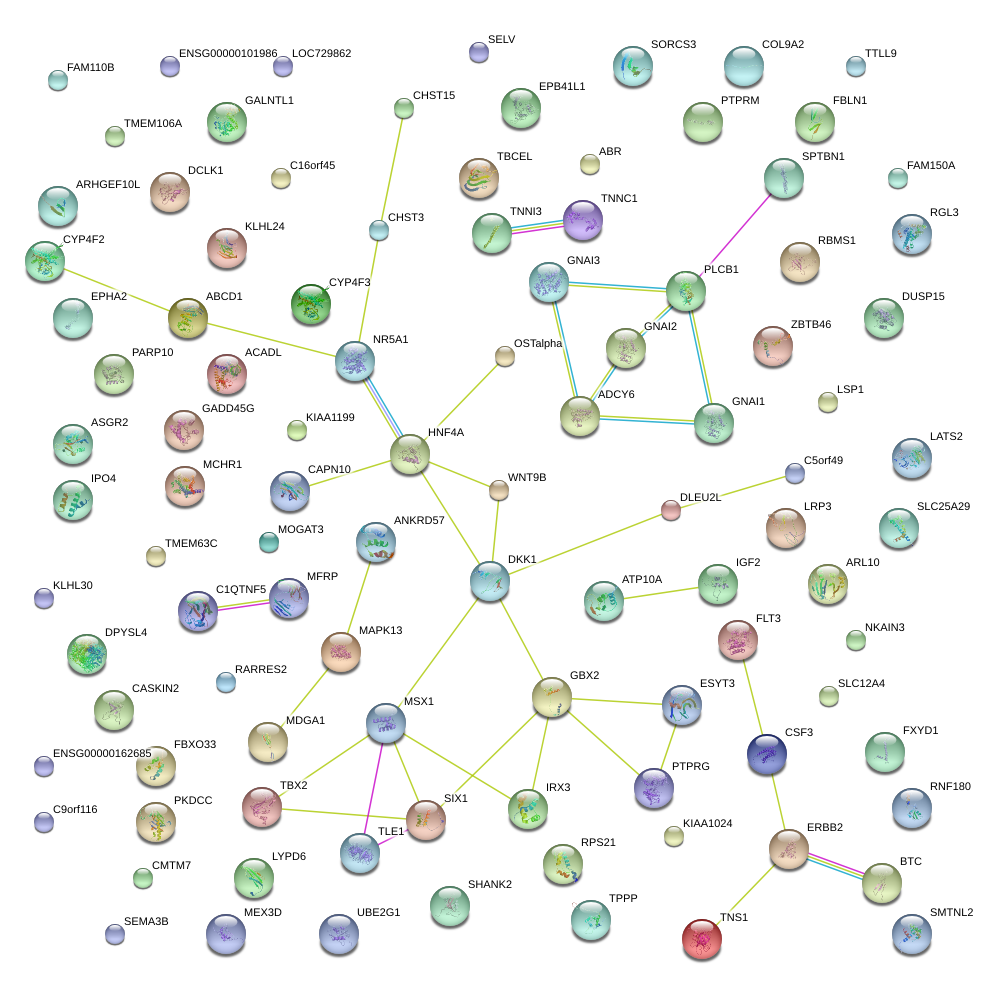
|
chr11_-_70641267
|
5.632
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr17_+_56832255
|
4.655
|
|
TBX2
|
T-box 2
|
|
chr2_+_238712101
|
4.472
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr20_+_8060762
|
4.429
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr17_+_56831951
|
4.422
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr1_-_16355013
|
3.838
|
NM_004431
|
EPHA2
|
EPH receptor A2
|
|
chr19_-_11390948
|
3.587
|
NM_001035223
NM_001161616
|
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3
|
|
chr11_-_2116048
|
3.444
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_-_959007
|
3.395
|
NM_001092
|
ABR
|
active BCR-related gene
|
|
chr17_+_56832492
|
3.287
|
|
TBX2
|
T-box 2
|
|
chr19_+_38377329
|
3.225
|
NM_002333
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr3_-_52463070
|
3.069
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chr6_+_36206239
|
2.995
|
NM_002754
|
MAPK13
|
mitogen-activated protein kinase 13
|
|
chr5_+_63497408
|
2.886
|
NM_001113561
NM_178532
|
RNF180
|
ring finger protein 180
|
|
chr19_+_38376980
|
2.886
|
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr11_-_2116360
|
2.812
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr2_-_210798232
|
2.786
|
NM_001608
|
ACADL
|
acyl-CoA dehydrogenase, long chain
|
|
chr3_+_61522922
|
2.703
|
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr14_-_38971049
|
2.697
|
|
FBXO33
|
F-box protein 33
|
|
chrX_+_152644385
|
2.691
|
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr10_+_106391032
|
2.623
|
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr7_+_79602023
|
2.592
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chr10_+_53744043
|
2.582
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr19_-_1518875
|
2.456
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr8_-_53640520
|
2.449
|
NM_207413
|
FAM150A
|
family with sequence similarity 150, member A
|
|
chr19_+_15612706
|
2.410
|
NM_000896
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr17_-_71023048
|
2.383
|
|
CASKIN2
|
CASK interacting protein 2
|
|
chr2_+_54537842
|
2.382
|
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr8_+_63324053
|
2.382
|
NM_173688
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3
|
|
chr17_-_71023221
|
2.372
|
NM_020753
|
CASKIN2
|
CASK interacting protein 2
|
|
chr5_-_7904255
|
2.359
|
NM_001089584
|
C5orf49
|
chromosome 5 open reading frame 49
|
|
chr14_-_99842519
|
2.345
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr14_+_68796621
|
2.338
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr20_+_42417752
|
2.330
|
NM_001030003
NM_001030004
NM_175914
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr2_-_218516950
|
2.302
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr18_+_7557313
|
2.215
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr3_+_184836049
|
2.190
|
NM_017644
|
KLHL24
|
kelch-like 24 (Drosophila)
|
|
chr17_+_42283946
|
2.188
|
NM_003396
|
WNT9B
|
wingless-type MMTV integration site family, member 9B
|
|
chr11_+_2116486
|
2.153
|
|
|
|
|
chr2_+_42128521
|
2.150
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr19_-_15869870
|
2.145
|
NM_001082
|
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2
|
|
chr3_+_139636104
|
2.134
|
NM_031913
|
ESYT3
|
extended synaptotagmin-like protein 3
|
|
chr11_-_2116501
|
2.112
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr15_+_78858763
|
2.075
|
NM_018689
|
KIAA1199
|
KIAA1199
|
|
chr4_+_4912272
|
2.071
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr3_+_139636111
|
2.063
|
|
ESYT3
|
extended synaptotagmin-like protein 3
|
|
chr11_+_1848674
|
2.060
|
NM_001013254
NM_001013255
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr20_-_29921461
|
1.972
|
NM_001012644
|
DUSP15
|
dual specificity phosphatase 15
|
|
chr15_-_23659329
|
1.967
|
NM_024490
|
ATP10A
|
ATPase, class V, type 10A
|
|
chr19_+_44697592
|
1.957
|
NM_182704
|
SELV
|
selenoprotein V
|
|
chr14_-_60185659
|
1.955
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr11_+_120399966
|
1.947
|
NM_001130047
NM_152715
|
TBCEL
|
tubulin folding cofactor E-like
|
|
chr20_-_29921983
|
1.933
|
NM_080611
|
DUSP15
C20orf57
|
dual specificity phosphatase 15
chromosome 20 open reading frame 57
|
|
chr17_-_37221443
|
1.911
|
|
|
|
|
chr1_-_40555413
|
1.910
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr11_-_118716736
|
1.905
|
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr20_-_29921671
|
1.885
|
NM_177991
|
DUSP15
|
dual specificity phosphatase 15
|
|
chr17_+_4434582
|
1.866
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr4_-_75938710
|
1.859
|
NM_001729
|
BTC
|
betacellulin
|
|
chr16_+_15435825
|
1.849
|
NM_033201
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr22_+_44277354
|
1.845
|
NM_001996
NM_006485
NM_006486
NM_006487
|
FBLN1
|
fibulin 1
|
|
chr7_+_78920846
|
1.845
|
|
LOC100505881
|
hypothetical LOC100505881
|
|
chr2_-_161058550
|
1.822
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr10_-_125841840
|
1.817
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr22_+_39404940
|
1.811
|
NM_005297
|
MCHR1
|
melanin-concentrating hormone receptor 1
|
|
chr19_+_40322231
|
1.803
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr8_+_59069522
|
1.789
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr3_+_121296290
|
1.789
|
|
|
|
|
chr9_+_91409746
|
1.780
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr20_+_29922165
|
1.777
|
NM_001008409
|
TTLL9
|
tubulin tyrosine ligase-like family, member 9
|
|
chr20_+_60395515
|
1.774
|
NM_001024
|
RPS21
|
ribosomal protein S21
|
|
chr3_+_197427779
|
1.762
|
NM_152672
|
OSTalpha
|
organic solute transporter alpha
|
|
chr3_+_32408346
|
1.754
|
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7
|
|
chr17_-_6958773
|
1.740
|
NM_001181
NM_080912
NM_080913
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr14_+_76717880
|
1.726
|
|
TMEM63C
|
transmembrane protein 63C
|
|
chr10_+_73393998
|
1.723
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr9_+_8848130
|
1.712
|
|
|
|
|
chr9_-_83494198
|
1.700
|
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr12_-_47468985
|
1.697
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr2_+_109729176
|
1.694
|
NM_023016
|
ANKRD57
|
ankyrin repeat domain 57
|
|
chr13_-_20533629
|
1.688
|
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr2_+_149895274
|
1.678
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr9_-_126309510
|
1.672
|
NM_004959
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr17_+_35425139
|
1.668
|
NM_000759
NM_001178147
NM_172219
NM_172220
|
CSF3
|
colony stimulating factor 3 (granulocyte)
|
|
chr19_-_38485161
|
1.658
|
|
|
|
|
chr6_-_37773609
|
1.657
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr7_-_149669622
|
1.654
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr15_+_77511912
|
1.652
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr13_-_35603466
|
1.650
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr20_+_34206070
|
1.646
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr13_-_49597776
|
1.642
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr10_+_133850325
|
1.642
|
NM_006426
|
DPYSL4
|
dihydropyrimidinase-like 4
|
|
chr16_-_52876390
|
1.631
|
|
IRX3
|
iroquois homeobox 3
|
|
chr9_-_137531517
|
1.626
|
NM_001048265
NM_144654
|
C9orf116
|
chromosome 9 open reading frame 116
|
|
chr14_-_23727826
|
1.617
|
|
IPO4
|
importin 4
|
|
chr8_-_145132551
|
1.606
|
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr17_-_6958695
|
1.593
|
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr17_+_38719371
|
1.587
|
NM_145041
|
TMEM106A
FLJ77644
|
transmembrane protein 106A
hypothetical LOC728772
|
|
chr17_+_35109802
|
1.586
|
|
ERBB2
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)
|
|
chr20_-_61933012
|
1.585
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr20_+_60395623
|
1.577
|
|
RPS21
|
ribosomal protein S21
|
|
chr3_+_50281582
|
1.574
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr2_-_236741352
|
1.573
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr17_-_4216359
|
1.566
|
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast)
|
|
chr1_+_17779528
|
1.565
|
NM_001011722
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr7_-_100630912
|
1.548
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr2_+_241174803
|
1.539
|
NM_023083
NM_023085
|
CAPN10
|
calpain 10
|
|
chr5_+_175725050
|
1.539
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr10_+_106390848
|
1.535
|
NM_014978
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr16_-_66535923
|
1.528
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr13_-_27572646
|
1.520
|
NM_004119
|
FLT3
|
fms-related tyrosine kinase 3
|
|
chr20_+_42417502
|
1.518
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr5_-_746413
|
1.517
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr2_+_9532087
|
1.514
|
NM_001039613
|
IAH1
|
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae)
|
|
chr15_+_66658626
|
1.488
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr14_-_23727850
|
1.488
|
NM_024658
|
IPO4
|
importin 4
|
|
chr2_-_42873179
|
1.487
|
NM_012205
|
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr8_-_145613058
|
1.485
|
NM_130849
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr2_-_73373956
|
1.477
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr7_-_78920806
|
1.459
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr16_+_66121040
|
1.457
|
NM_001193523
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr17_+_43403466
|
1.455
|
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr3_+_127725851
|
1.451
|
NM_152889
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr18_+_18969724
|
1.443
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr20_+_44771532
|
1.432
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr1_+_78126787
|
1.431
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr14_-_20562962
|
1.430
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr17_+_77574476
|
1.429
|
NM_144999
|
LRRC45
|
leucine rich repeat containing 45
|
|
chr5_+_146594718
|
1.427
|
NM_001112724
NM_145001
|
STK32A
|
serine/threonine kinase 32A
|
|
chr2_-_20288108
|
1.426
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr11_+_76455574
|
1.420
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chrX_+_66683075
|
1.417
|
|
AR
|
androgen receptor
|
|
chr1_-_925332
|
1.414
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr7_+_45894480
|
1.411
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr8_+_16929116
|
1.411
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr5_-_115938305
|
1.410
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr16_-_66535457
|
1.409
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chrX_+_53095230
|
1.405
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr16_-_69030421
|
1.396
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chrX_+_152565849
|
1.393
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr2_-_128792515
|
1.389
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr5_+_76542461
|
1.385
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr11_+_112337411
|
1.379
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr22_+_33792125
|
1.374
|
NM_001008494
|
ISX
|
intestine-specific homeobox
|
|
chr14_+_104218487
|
1.372
|
|
|
|
|
chr10_+_124210980
|
1.370
|
NM_002775
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr20_-_17610704
|
1.368
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr5_+_179166608
|
1.363
|
NM_001142299
|
SQSTM1
|
sequestosome 1
|
|
chr8_-_145099963
|
1.362
|
NM_201379
|
PLEC
|
plectin
|
|
chr3_-_10522267
|
1.351
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr2_+_242289983
|
1.345
|
NM_032329
|
ING5
|
inhibitor of growth family, member 5
|
|
chr2_+_241193492
|
1.336
|
NM_001195381
NM_001195382
|
GPR35
|
G protein-coupled receptor 35
|
|
chr22_-_22970850
|
1.328
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr5_-_179166378
|
1.326
|
NM_014275
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr5_-_37285160
|
1.323
|
NM_023073
|
C5orf42
|
chromosome 5 open reading frame 42
|
|
chr11_-_2116775
|
1.318
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr13_-_20533703
|
1.313
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr7_-_99594265
|
1.313
|
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr5_-_115180221
|
1.313
|
NM_001801
|
CDO1
|
cysteine dioxygenase, type I
|
|
chr14_-_38971370
|
1.311
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr17_+_75507750
|
1.311
|
|
|
|
|
chr17_-_71535076
|
1.311
|
NM_001988
|
EVPL
|
envoplakin
|
|
chr17_+_1129706
|
1.310
|
NM_172367
|
TUSC5
|
tumor suppressor candidate 5
|
|
chr5_-_175989930
|
1.307
|
NM_001001502
NM_003085
|
SNCB
|
synuclein, beta
|
|
chr10_-_105604942
|
1.306
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr11_+_110675175
|
1.304
|
NM_001136105
|
C11orf93
|
chromosome 11 open reading frame 93
|
|
chr16_-_66251611
|
1.304
|
|
ACD
|
adrenocortical dysplasia homolog (mouse)
|
|
chr21_+_41610530
|
1.303
|
NM_058186
NM_206964
|
FAM3B
|
family with sequence similarity 3, member B
|
|
chr16_-_21439195
|
1.302
|
|
SLC7A5P2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 pseudogene 2
|
|
chr2_-_161058120
|
1.301
|
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr2_-_219633481
|
1.299
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chrX_-_99551926
|
1.295
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chrX_+_128942435
|
1.295
|
|
|
|
|
chr17_-_37275142
|
1.289
|
NM_018143
|
KLHL11
|
kelch-like 11 (Drosophila)
|
|
chr16_+_84202515
|
1.288
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr10_-_131652007
|
1.288
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr20_+_61337679
|
1.286
|
NM_022161
NM_139317
|
BIRC7
|
baculoviral IAP repeat containing 7
|
|
chr13_-_52211925
|
1.282
|
NM_001011705
NM_007015
|
LECT1
|
leukocyte cell derived chemotaxin 1
|
|
chr7_+_882704
|
1.281
|
NM_015949
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae)
|
|
chr3_+_160964574
|
1.280
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr11_+_116205830
|
1.268
|
NM_000040
|
APOC3
|
apolipoprotein C-III
|
|
chr14_-_106282128
|
1.268
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr17_-_17340219
|
1.268
|
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr16_-_69277355
|
1.267
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr11_-_118799452
|
1.265
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr11_-_900793
|
1.262
|
|
CHID1
|
chitinase domain containing 1
|
|
chr4_+_8322358
|
1.262
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr9_-_35062643
|
1.261
|
NM_007126
|
VCP
|
valosin containing protein
|
|
chr3_+_134947940
|
1.258
|
|
TF
|
transferrin
|
|
chr9_-_119217147
|
1.257
|
|
ASTN2
|
astrotactin 2
|
|
chr9_-_116190038
|
1.256
|
|
AKNA
|
AT-hook transcription factor
|
|
chr8_-_22044463
|
1.254
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr3_+_134947573
|
1.253
|
NM_001063
|
TF
|
transferrin
|
|
chrX_+_68641802
|
1.253
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr3_-_198240969
|
1.244
|
|
MFI2
|
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5
|
|
chr1_+_1256585
|
1.242
|
NM_152228
|
TAS1R3
|
taste receptor, type 1, member 3
|
|
chr14_+_68796433
|
1.239
|
NM_001168368
NM_020692
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr7_+_17304707
|
1.238
|
NM_001621
|
AHR
|
aryl hydrocarbon receptor
|
|
chr17_+_43403475
|
1.237
|
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|
|
chr17_+_43403405
|
1.235
|
NM_176096
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3
|