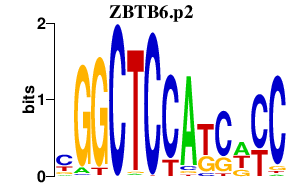
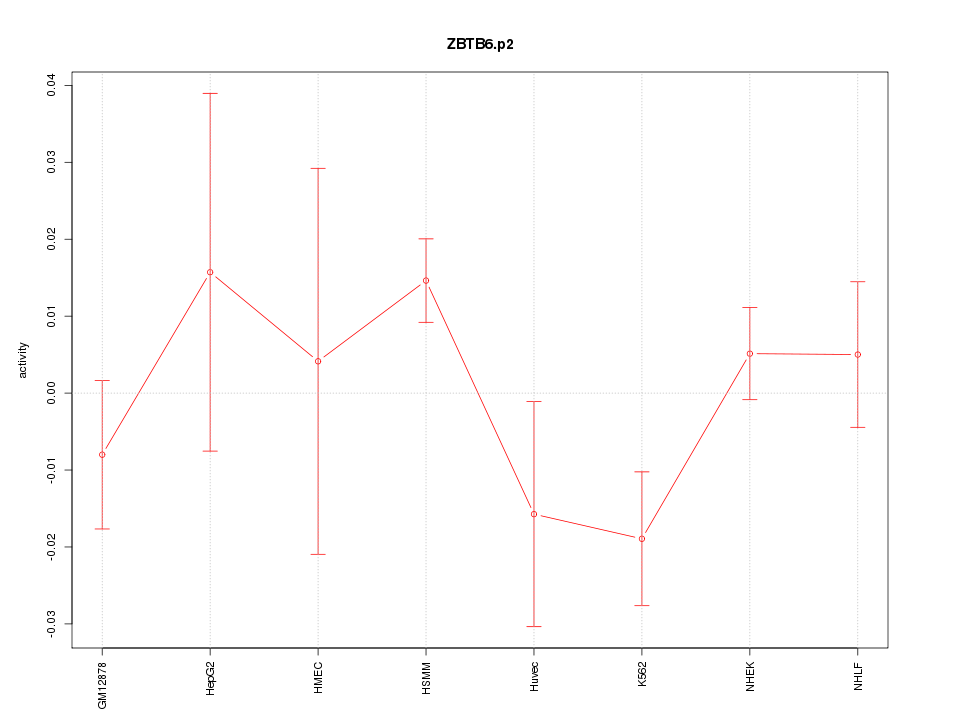
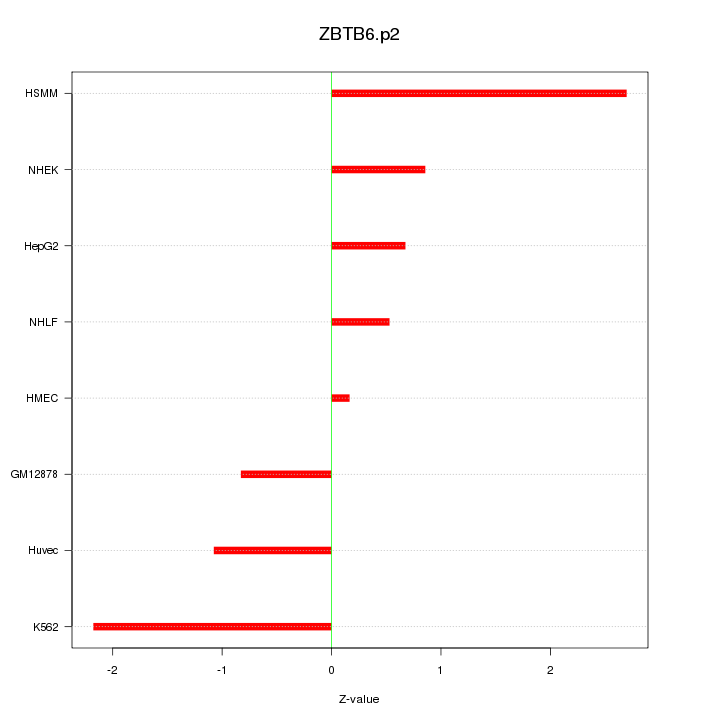
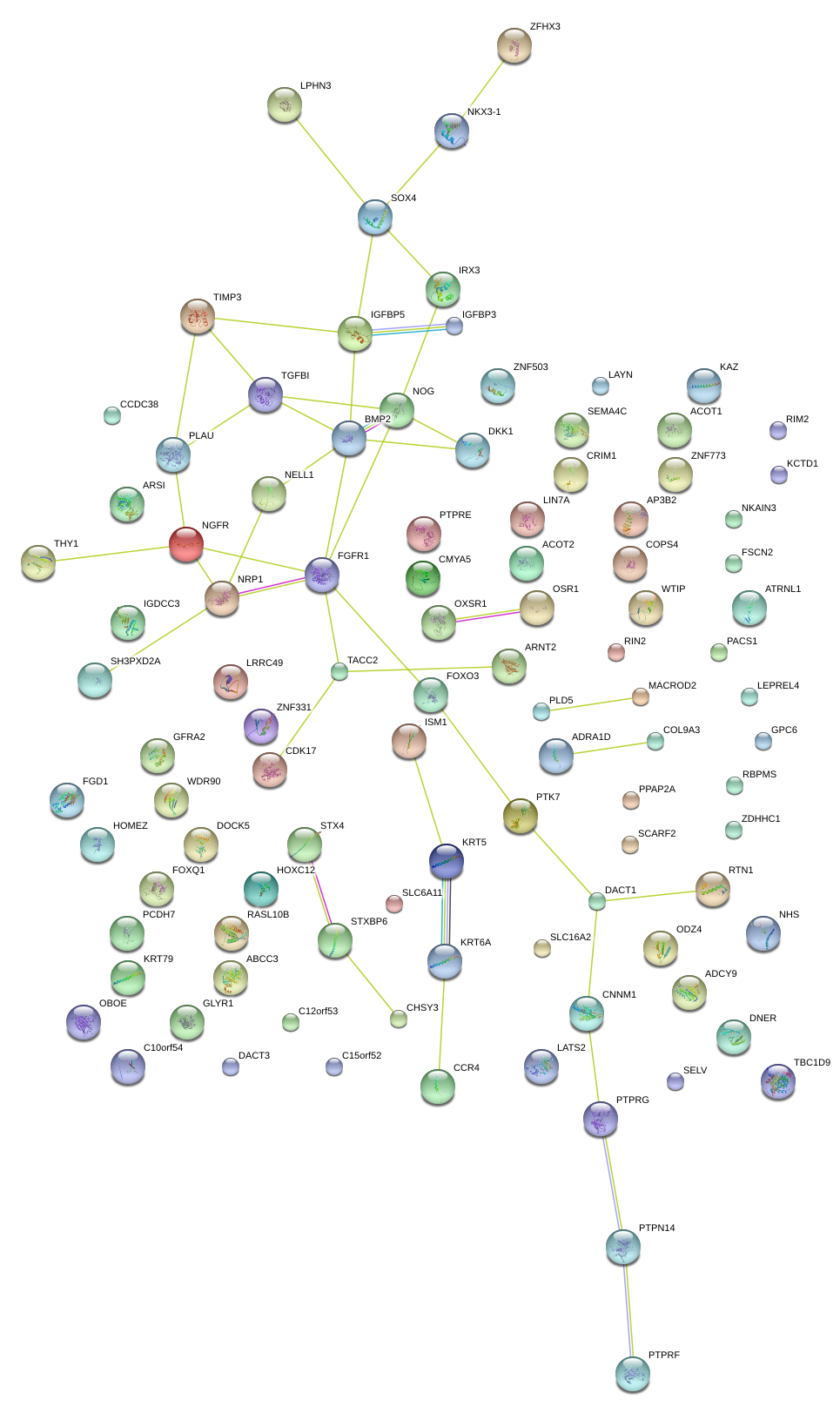
|
chr10_-_105604942
|
2.586
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr11_-_78829342
|
2.370
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr17_+_52026058
|
2.215
|
NM_005450
|
NOG
|
noggin
|
|
chr19_+_62703136
|
2.105
|
|
ZNF773
|
zinc finger protein 773
|
|
chr19_+_62703113
|
1.993
|
NM_198542
|
ZNF773
|
zinc finger protein 773
|
|
chr8_+_63324053
|
1.847
|
NM_173688
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3
|
|
chr16_-_71650034
|
1.636
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr22_+_31527678
|
1.541
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr12_-_79855580
|
1.537
|
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr22_+_31527744
|
1.499
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr16_-_66007650
|
1.430
|
NM_013304
|
ZDHHC1
|
zinc finger, DHHC-type containing 1
|
|
chr20_+_13150417
|
1.423
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr12_-_79855618
|
1.421
|
NM_004664
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr20_-_4177519
|
1.421
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr1_+_43769065
|
1.356
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr12_-_94860504
|
1.308
|
NM_182496
|
CCDC38
|
coiled-coil domain containing 38
|
|
chr10_+_101079099
|
1.298
|
|
CNNM1
|
cyclin M1
|
|
chr14_+_58174489
|
1.276
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr16_+_65018316
|
1.239
|
NM_001136106
NM_001178020
NM_001197224
NM_001197225
|
BEAN1
|
brain expressed, associated with NEDD4, 1
|
|
chr10_+_101078784
|
1.158
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chr16_-_52876390
|
1.154
|
|
IRX3
|
iroquois homeobox 3
|
|
chr17_+_31082791
|
1.112
|
NM_033315
|
RASL10B
|
RAS-like, family 10, member B
|
|
chr13_+_92677012
|
1.090
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr8_+_104582151
|
1.081
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr12_-_51200347
|
1.070
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr19_+_38485583
|
1.040
|
|
LOC80054
|
hypothetical LOC80054
|
|
chr18_-_22382497
|
1.040
|
NM_001142730
|
KCTD1
|
potassium channel tetramerisation domain containing 1
|
|
chr6_+_1257674
|
1.029
|
NM_033260
|
FOXQ1
|
forkhead box Q1
|
|
chr1_-_240754620
|
1.027
|
NM_152666
|
PLD5
|
phospholipase D family, member 5
|
|
chr10_-_76829857
|
1.011
|
|
ZNF503
|
zinc finger protein 503
|
|
chr5_-_149662640
|
1.010
|
NM_001012301
|
ARSI
|
arylsulfatase family, member I
|
|
chr3_+_10832867
|
1.007
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr14_-_24588916
|
1.005
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr5_+_79021401
|
0.999
|
NM_153610
|
CMYA5
|
cardiomyopathy associated 5
|
|
chr16_+_639278
|
0.984
|
NM_145294
|
WDR90
|
WD repeat domain 90
|
|
chr8_+_30361485
|
0.957
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr13_-_20533703
|
0.955
|
NM_014572
|
LATS2
|
LATS, large tumor suppressor, homolog 2 (Drosophila)
|
|
chr17_+_44927591
|
0.946
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr10_+_75343352
|
0.945
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr1_-_212791188
|
0.940
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr1_-_212791467
|
0.926
|
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr14_-_59407103
|
0.918
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr15_-_63457378
|
0.910
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chr2_-_19421852
|
0.907
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr20_+_13924014
|
0.902
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chrX_+_17303798
|
0.891
|
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr20_+_6696744
|
0.889
|
NM_001200
|
BMP2
|
bone morphogenetic protein 2
|
|
chrX_+_73557809
|
0.882
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr14_+_73073570
|
0.864
|
NM_001037161
|
ACOT1
|
acyl-CoA thioesterase 1
|
|
chr2_-_230287442
|
0.847
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr5_-_54866126
|
0.841
|
NM_003711
NM_176895
|
RNF138P1
PPAP2A
|
ring finger protein 138 pseudogene 1
phosphatidic acid phosphatase type 2A
|
|
chr15_+_78483624
|
0.837
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr6_+_19946591
|
0.813
|
|
|
|
|
chr15_-_38420414
|
0.813
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr2_+_36436873
|
0.810
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr10_+_123862430
|
0.807
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr10_-_33663984
|
0.802
|
|
NRP1
|
neuropilin 1
|
|
chr11_-_118799058
|
0.788
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr6_+_108988761
|
0.784
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr16_+_30952382
|
0.784
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr16_-_4105703
|
0.782
|
|
ADCY9
|
adenylate cyclase 9
|
|
chrX_+_17303458
|
0.776
|
NM_198270
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr4_+_30331543
|
0.766
|
|
PCDH7
|
protocadherin 7
|
|
chr19_+_44697592
|
0.762
|
NM_182704
|
SELV
|
selenoprotein V
|
|
chr6_+_43151922
|
0.760
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr19_+_58750305
|
0.754
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr8_-_21702273
|
0.753
|
NM_001165038
NM_001165039
NM_001495
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr17_-_37221976
|
0.748
|
NM_006455
|
LEPREL4
|
leprecan-like 4
|
|
chr10_+_116843108
|
0.748
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr4_-_141896920
|
0.746
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr17_+_77110011
|
0.743
|
NM_001077182
NM_012418
|
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus)
|
|
chr6_+_21701942
|
0.739
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr22_-_19122047
|
0.738
|
|
SCARF2
|
scavenger receptor class F, member 2
|
|
chr12_+_52634950
|
0.736
|
NM_173860
|
HOXC12
|
homeobox C12
|
|
chr17_+_46067114
|
0.733
|
NM_001144070
NM_003786
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3
|
|
chrX_-_54538591
|
0.731
|
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1
|
|
chr5_+_135392482
|
0.729
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr8_-_38444982
|
0.725
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr20_+_19903778
|
0.719
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chrX_+_17303727
|
0.718
|
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies)
|
|
chr16_+_30952350
|
0.714
|
|
STX4
|
syntaxin 4
|
|
chr5_+_135392624
|
0.709
|
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr2_-_217267742
|
0.705
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr8_-_23596346
|
0.702
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr4_+_61749540
|
0.701
|
|
LPHN3
|
latrophilin 3
|
|
chr19_+_39664706
|
0.697
|
NM_001080436
|
WTIP
|
Wilms tumor 1 interacting protein
|
|
chr11_+_110916629
|
0.694
|
|
LAYN
|
layilin
|
|
chr10_-_73203202
|
0.686
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr4_+_30330963
|
0.686
|
NM_001173523
NM_002589
NM_032456
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr16_-_4837266
|
0.684
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr10_+_129595289
|
0.682
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr15_+_68971835
|
0.682
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr2_-_96899396
|
0.681
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr12_-_6680053
|
0.678
|
NM_153685
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
chr8_+_25098296
|
0.665
|
|
DOCK5
|
dedicator of cytokinesis 5
|
|
chr20_+_13924263
|
0.664
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr3_+_61522310
|
0.663
|
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr15_-_81175542
|
0.659
|
NM_004644
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr10_+_53744043
|
0.652
|
NM_012242
|
DKK1
|
dickkopf homolog 1 (Xenopus laevis)
|
|
chr12_-_95317928
|
0.652
|
|
CDK17
|
cyclin-dependent kinase 17
|
|
chr10_-_33663777
|
0.644
|
NM_001024628
NM_001024629
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr5_+_129268352
|
0.640
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr20_+_60918836
|
0.637
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr7_-_45927263
|
0.634
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr20_+_2581177
|
0.633
|
NM_006392
|
NOP56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr3_-_49736297
|
0.633
|
NM_013334
NM_021971
|
GMPPB
|
GDP-mannose pyrophosphorylase B
|
|
chr1_-_212791597
|
0.631
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr12_-_105165805
|
0.630
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr19_+_627346
|
0.630
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr11_+_110916442
|
0.629
|
NM_178834
|
LAYN
|
layilin
|
|
chr2_+_39746525
|
0.627
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr11_-_75057062
|
0.625
|
NM_033063
NM_207577
|
MAP6
|
microtubule-associated protein 6
|
|
chr2_+_190916440
|
0.624
|
NM_001128928
NM_002194
|
INPP1
|
inositol polyphosphate-1-phosphatase
|
|
chr11_-_118799452
|
0.620
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr2_-_86889029
|
0.615
|
NM_001145873
|
CD8A
|
CD8a molecule
|
|
chr6_+_1555679
|
0.615
|
NM_001453
|
FOXC1
|
forkhead box C1
|
|
chr4_-_75938710
|
0.614
|
NM_001729
|
BTC
|
betacellulin
|
|
chr16_+_67698943
|
0.614
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr3_+_112273464
|
0.611
|
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr3_-_191522886
|
0.606
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr8_-_38444398
|
0.606
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr22_+_48740432
|
0.604
|
|
PIM3
|
pim-3 oncogene
|
|
chr11_-_75056761
|
0.603
|
|
MAP6
|
microtubule-associated protein 6
|
|
chr12_-_51173238
|
0.598
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr5_-_57791544
|
0.594
|
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr3_+_112273352
|
0.593
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr12_-_6354650
|
0.592
|
NM_001159576
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr17_-_39437362
|
0.591
|
NM_004160
|
PYY
|
peptide YY
|
|
chr1_+_149221122
|
0.590
|
NM_003568
|
ANXA9
|
annexin A9
|
|
chr5_-_58370693
|
0.588
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr6_+_45497796
|
0.588
|
NM_004348
|
RUNX2
|
runt-related transcription factor 2
|
|
chr16_-_52877467
|
0.586
|
|
IRX3
|
iroquois homeobox 3
|
|
chr11_+_116575249
|
0.585
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr17_-_45427565
|
0.584
|
NM_005220
|
DLX3
|
distal-less homeobox 3
|
|
chr17_-_1478293
|
0.583
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr15_+_72006015
|
0.583
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_+_22751162
|
0.579
|
NM_003097
NM_022804
NM_005678
|
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N
SNRPN upstream reading frame
|
|
chr4_+_30331289
|
0.578
|
|
PCDH7
|
protocadherin 7
|
|
chr16_+_55217085
|
0.578
|
NM_175617
|
MT1E
|
metallothionein 1E
|
|
chr1_+_13782838
|
0.577
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr4_+_1764802
|
0.577
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr4_-_111777956
|
0.575
|
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr19_-_50977586
|
0.571
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr8_+_63323739
|
0.569
|
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3
|
|
chr10_-_125843092
|
0.566
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr19_+_62690890
|
0.566
|
NM_001098491
NM_001098492
NM_001098493
NM_001098494
NM_001098495
NM_001098496
NM_024691
|
ZNF419
|
zinc finger protein 419
|
|
chr12_+_131774225
|
0.565
|
NM_018663
|
PXMP2
|
peroxisomal membrane protein 2, 22kDa
|
|
chr17_-_7323667
|
0.563
|
NM_001128833
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr1_+_156417302
|
0.561
|
|
CD1D
|
CD1d molecule
|
|
chr5_+_178300799
|
0.561
|
NM_001178089
NM_001178090
NM_182594
|
ZNF454
|
zinc finger protein 454
|
|
chr1_-_20684858
|
0.559
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr7_-_19123765
|
0.554
|
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr3_-_27500882
|
0.553
|
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr18_+_7557313
|
0.549
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr10_+_104619199
|
0.547
|
NM_020682
|
AS3MT
|
arsenic (+3 oxidation state) methyltransferase
|
|
chrX_-_131451675
|
0.543
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chrX_-_50230329
|
0.543
|
NM_001013742
|
DGKK
|
diacylglycerol kinase, kappa
|
|
chr1_-_59020875
|
0.536
|
|
JUN
|
jun proto-oncogene
|
|
chr10_+_96152262
|
0.535
|
|
TBC1D12
|
TBC1 domain family, member 12
|
|
chr9_-_85342868
|
0.535
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr16_-_28411123
|
0.535
|
NM_001042432
|
CLN3
|
ceroid-lipofuscinosis, neuronal 3
|
|
chr16_+_48657381
|
0.533
|
NM_182922
|
HEATR3
|
HEAT repeat containing 3
|
|
chr12_-_6354818
|
0.532
|
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr1_+_201363458
|
0.531
|
NM_000674
NM_001048230
|
ADORA1
|
adenosine A1 receptor
|
|
chr18_-_24011349
|
0.530
|
NM_001792
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal)
|
|
chr11_-_35503720
|
0.529
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr6_+_30335314
|
0.528
|
|
HLA-L
|
major histocompatibility complex, class I, L, pseudogene
|
|
chr7_-_19123787
|
0.524
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr16_+_29735208
|
0.523
|
|
C16orf53
|
chromosome 16 open reading frame 53
|
|
chr20_+_29565898
|
0.520
|
NM_030789
NM_178580
NM_178581
NM_178582
|
HM13
|
histocompatibility (minor) 13
|
|
chr1_+_64012154
|
0.520
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr4_-_17392232
|
0.520
|
NM_015688
|
FAM184B
|
family with sequence similarity 184, member B
|
|
chr18_+_40513889
|
0.518
|
NM_001130110
|
SETBP1
|
SET binding protein 1
|
|
chr9_-_21325239
|
0.518
|
NM_018847
|
KLHL9
|
kelch-like 9 (Drosophila)
|
|
chr20_-_9767406
|
0.517
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr17_+_39437439
|
0.516
|
NM_153006
|
NAGS
|
N-acetylglutamate synthase
|
|
chr13_+_27392136
|
0.516
|
NM_000209
|
PDX1
|
pancreatic and duodenal homeobox 1
|
|
chr5_+_167889123
|
0.515
|
|
|
|
|
chr20_+_29565922
|
0.514
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr2_+_85213884
|
0.510
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr20_-_3166834
|
0.504
|
NM_001174090
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11
|
|
chr16_+_13921490
|
0.504
|
NM_005236
|
ERCC4
|
excision repair cross-complementing rodent repair deficiency, complementation group 4
|
|
chr10_-_99780574
|
0.503
|
NM_018058
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr3_-_75566781
|
0.501
|
|
FAM86DP
|
family with sequence similarity 86, member D, pseudogene
|
|
chr3_-_38046034
|
0.501
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr6_-_131426014
|
0.499
|
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2
|
|
chr22_-_45512794
|
0.496
|
NM_022766
|
CERK
|
ceramide kinase
|
|
chr4_+_52612197
|
0.496
|
NM_145263
|
SPATA18
|
spermatogenesis associated 18 homolog (rat)
|
|
chr11_+_129823667
|
0.495
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr2_-_65512619
|
0.494
|
|
SPRED2
|
sprouty-related, EVH1 domain containing 2
|
|
chr17_-_76985538
|
0.491
|
|
|
|
|
chr3_-_48675321
|
0.491
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr11_-_73149705
|
0.491
|
NM_002869
NM_198896
|
RAB6A
|
RAB6A, member RAS oncogene family
|
|
chrX_-_92815222
|
0.489
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr2_-_42873179
|
0.489
|
NM_012205
|
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr15_-_30798417
|
0.489
|
|
|
|
|
chr9_-_122731263
|
0.488
|
NM_001190945
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr10_+_103815114
|
0.485
|
NM_024747
|
HPS6
|
Hermansky-Pudlak syndrome 6
|
|
chr9_+_34948320
|
0.484
|
|
KIAA1045
|
KIAA1045
|
|
chr6_-_80713589
|
0.482
|
NM_022726
|
ELOVL4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|