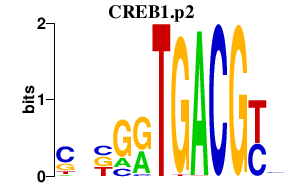
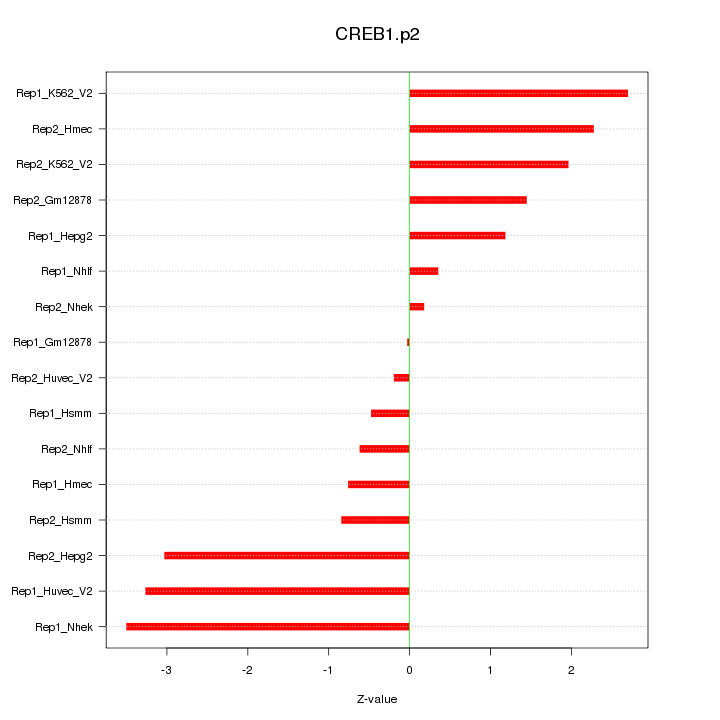
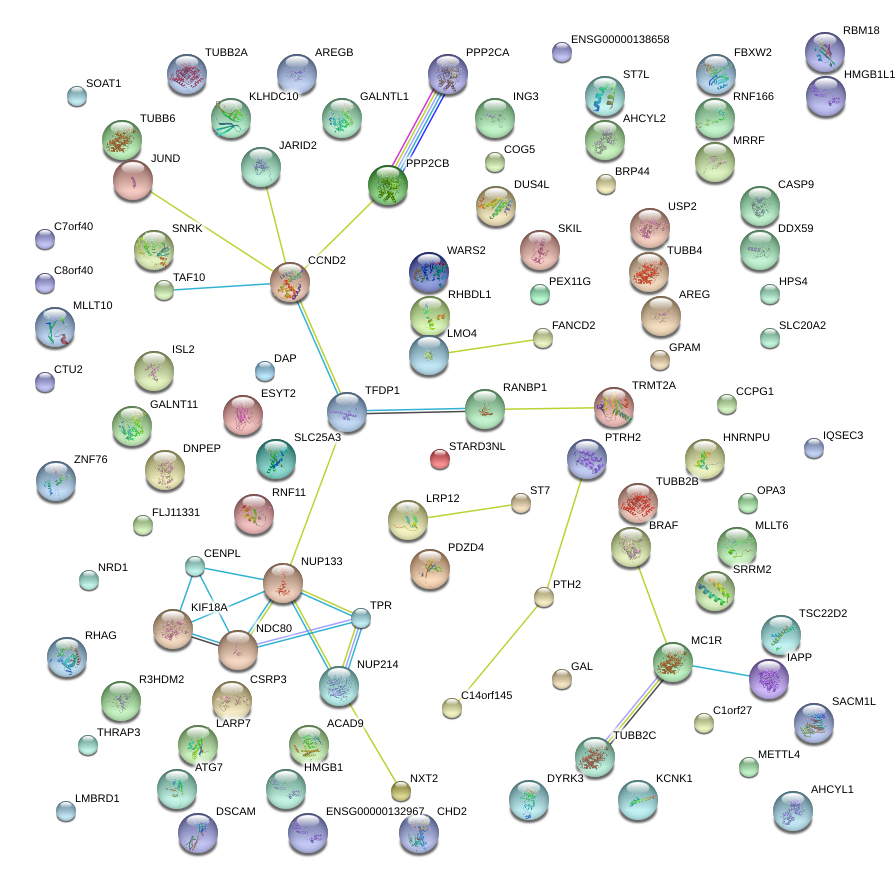
|
chr1_-_15723279
|
2.264
|
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr1_-_15723305
|
2.026
|
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr1_-_119484733
|
1.991
|
NM_015836
NM_201263
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial
|
|
chr1_-_243092581
|
1.855
|
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr1_+_119484529
|
1.821
|
|
|
|
|
chr7_-_158314963
|
1.601
|
NM_020728
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr4_-_113777569
|
1.570
|
NM_018392
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr6_-_15356865
|
1.563
|
|
|
|
|
chr4_+_113777568
|
1.557
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr9_+_124066973
|
1.543
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr4_-_113777470
|
1.543
|
|
C4orf21
|
chromosome 4 open reading frame 21
|
|
chr7_-_140270749
|
1.541
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr16_+_2742709
|
1.486
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr9_-_124066901
|
1.466
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr13_+_113287048
|
1.446
|
NM_007111
|
TFDP1
|
transcription factor Dp-1
|
|
chr15_+_74416129
|
1.445
|
|
ISL2
|
ISL LIM homeobox 2
|
|
chr1_-_15723363
|
1.440
|
NM_001229
NM_032996
|
CASP9
|
caspase 9, apoptosis-related cysteine peptidase
|
|
chr9_-_124066838
|
1.433
|
|
RBM18
|
RNA binding motif protein 18
|
|
chr15_+_74416156
|
1.393
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr7_-_44992729
|
1.382
|
|
C7orf40
|
chromosome 7 open reading frame 40
|
|
chr15_-_53487748
|
1.290
|
NM_004748
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chr18_+_2561509
|
1.288
|
NM_006101
|
NDC80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr3_+_171558215
|
1.264
|
|
SKIL
|
SKI-like oncogene
|
|
chr9_+_124066702
|
1.262
|
NM_001173512
NM_138777
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr6_-_70563193
|
1.248
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr3_+_171558188
|
1.239
|
|
SKIL
|
SKI-like oncogene
|
|
chr4_+_75529716
|
1.213
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr1_+_110328955
|
1.169
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr13_+_113286979
|
1.126
|
|
TFDP1
|
transcription factor Dp-1
|
|
chr22_+_18485145
|
1.123
|
|
RANBP1
|
RAN binding protein 1
|
|
chr12_-_56111054
|
1.119
|
|
R3HDM2
|
R3H domain containing 2
|
|
chr1_-_198905714
|
1.114
|
NM_001031725
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr18_-_2561422
|
1.108
|
NM_022840
|
METTL4
|
methyltransferase like 4
|
|
chr3_+_151608673
|
1.108
|
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr18_+_2561587
|
1.100
|
|
NDC80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr16_+_2742704
|
1.096
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr1_-_198905619
|
1.096
|
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr1_+_110328941
|
1.091
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr22_+_18485057
|
1.050
|
|
RANBP1
|
RAN binding protein 1
|
|
chr16_+_2742684
|
1.042
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr7_+_116447499
|
1.041
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr16_-_87300233
|
1.038
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr16_+_87300385
|
1.019
|
NM_001012759
NM_001012762
|
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe)
|
|
chr19_-_18253413
|
1.009
|
NM_005354
|
JUND
|
jun D proto-oncogene
|
|
chr16_-_87300308
|
0.978
|
NM_178841
|
RNF166
|
ring finger protein 166
|
|
chr19_-_6453224
|
0.963
|
NM_006087
|
TUBB4
|
tubulin, beta 4
|
|
chr11_-_118757477
|
0.961
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr6_+_15357013
|
0.955
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr3_+_10043139
|
0.943
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr7_+_151353707
|
0.942
|
NM_022087
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr1_+_110328850
|
0.932
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr12_+_97511805
|
0.932
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr22_+_18485076
|
0.929
|
|
RANBP1
|
RAN binding protein 1
|
|
chr6_-_70563245
|
0.924
|
|
LMBRD1
|
LMBR1 domain containing 1
|
|
chr7_+_151353759
|
0.923
|
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr4_+_75699652
|
0.920
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr11_-_118757574
|
0.917
|
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr12_+_46133
|
0.910
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr3_+_130081022
|
0.908
|
NM_014049
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr22_+_18485095
|
0.902
|
|
RANBP1
|
RAN binding protein 1
|
|
chr12_+_97511585
|
0.889
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr1_+_87567164
|
0.887
|
|
LMO4
|
LIM domain only 4
|
|
chr12_+_97511596
|
0.884
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr12_+_97511631
|
0.881
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr7_+_120378052
|
0.870
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr7_+_38184393
|
0.868
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr15_+_91248706
|
0.868
|
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr3_+_11288975
|
0.862
|
NM_001136031
NM_001144912
NM_006395
|
ATG7
|
ATG7 autophagy related 7 homolog (S. cerevisiae)
|
|
chr1_+_110329279
|
0.853
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr1_+_141560298
|
0.849
|
|
|
|
|
chr12_+_97511594
|
0.843
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr1_+_184611512
|
0.841
|
NM_001164245
NM_001164246
NM_017847
|
C1orf27
|
chromosome 1 open reading frame 27
|
|
chr9_+_132990834
|
0.836
|
|
NUP214
|
nucleoporin 214kDa
|
|
chr1_+_231816372
|
0.836
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr1_+_87567136
|
0.833
|
|
LMO4
|
LIM domain only 4
|
|
chr3_+_11289092
|
0.832
|
|
ATG7
|
ATG7 autophagy related 7 homolog (S. cerevisiae)
|
|
chr7_+_151353775
|
0.830
|
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11)
|
|
chr22_-_18484428
|
0.830
|
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr1_+_110328830
|
0.829
|
NM_006621
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr19_+_47593137
|
0.827
|
|
|
|
|
chr7_+_38184444
|
0.825
|
NM_032016
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr3_+_11289065
|
0.825
|
|
ATG7
|
ATG7 autophagy related 7 homolog (S. cerevisiae)
|
|
chr3_+_130081187
|
0.822
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr16_-_53520208
|
0.821
|
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr1_+_87566738
|
0.820
|
NM_006769
|
LMO4
|
LIM domain only 4
|
|
chr6_-_49712483
|
0.819
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr19_-_18253436
|
0.817
|
|
JUND
|
jun D proto-oncogene
|
|
chr1_-_172059780
|
0.815
|
NM_001171182
|
CENPL
|
centromere protein L
|
|
chr12_+_97511520
|
0.813
|
NM_002635
NM_005888
NM_213611
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr10_+_21863099
|
0.813
|
NM_001195627
NM_001195628
NM_001195630
NM_004641
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr11_-_28086263
|
0.810
|
|
KIF18A
|
kinesin family member 18A
|
|
chr7_+_38184375
|
0.796
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr7_+_129497572
|
0.792
|
NM_014997
|
KLHDC10
|
kelch domain containing 10
|
|
chr11_-_28086277
|
0.792
|
NM_031217
|
KIF18A
|
kinesin family member 18A
|
|
chrX_+_108666891
|
0.790
|
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr16_+_2742677
|
0.782
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr16_+_2742641
|
0.777
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr12_+_97511601
|
0.772
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr16_+_2742674
|
0.770
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr7_-_106991572
|
0.767
|
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr12_+_97511569
|
0.767
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr1_-_166171838
|
0.763
|
NM_015415
|
BRP44
|
brain protein 44
|
|
chr1_-_172060077
|
0.758
|
|
CENPL
|
centromere protein L
|
|
chr19_-_7459867
|
0.756
|
NM_080662
|
PEX11G
|
peroxisomal biogenesis factor 11 gamma
|
|
chr9_+_132990760
|
0.753
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr5_-_10814341
|
0.749
|
|
DAP
|
death-associated protein
|
|
chr1_-_166171702
|
0.746
|
|
BRP44
|
brain protein 44
|
|
chr8_-_42515774
|
0.745
|
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr22_-_18484695
|
0.744
|
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr19_-_50779924
|
0.741
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr6_-_31779033
|
0.738
|
|
ABHD16A
|
abhydrolase domain containing 16A
|
|
chr12_+_4253143
|
0.738
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr3_+_45705918
|
0.737
|
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast)
|
|
chr3_+_45705845
|
0.736
|
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast)
|
|
chr9_-_122595349
|
0.736
|
|
FBXW2
|
F-box and WD repeat domain containing 2
|
|
chr22_-_25209724
|
0.735
|
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr19_-_18253259
|
0.731
|
|
|
|
|
chr6_-_31779068
|
0.723
|
|
ABHD16A
|
abhydrolase domain containing 16A
|
|
chr7_+_129497637
|
0.722
|
|
KLHDC10
|
kelch domain containing 10
|
|
chr7_+_106991638
|
0.722
|
NM_181581
|
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae)
|
|
chr5_-_10814344
|
0.722
|
NM_004394
|
DAP
|
death-associated protein
|
|
chr22_+_18485068
|
0.718
|
|
RANBP1
|
RAN binding protein 1
|
|
chr6_+_35335456
|
0.715
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr3_+_10043097
|
0.704
|
NM_001018115
NM_033084
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr16_+_2742606
|
0.698
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr3_+_130081161
|
0.696
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr16_+_2742657
|
0.696
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr10_-_113933457
|
0.695
|
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr3_+_10043123
|
0.694
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr5_-_10814179
|
0.693
|
|
DAP
|
death-associated protein
|
|
chr5_-_133589450
|
0.692
|
|
PPP2CA
|
protein phosphatase 2, catalytic subunit, alpha isozyme
|
|
chr3_+_43303005
|
0.689
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr1_+_204875671
|
0.689
|
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr3_+_130081210
|
0.689
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr13_-_29938029
|
0.687
|
NM_002128
|
HMGB1
|
high-mobility group box 1
|
|
chr16_+_665680
|
0.686
|
|
RHBDL1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr16_+_2742635
|
0.685
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr1_-_184611424
|
0.682
|
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chr3_+_130081130
|
0.681
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr1_+_51474558
|
0.677
|
|
RNF11
|
ring finger protein 11
|
|
chr9_-_122595413
|
0.676
|
|
FBXW2
|
F-box and WD repeat domain containing 2
|
|
chr3_+_130081149
|
0.674
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr5_+_10406980
|
0.672
|
|
|
|
|
chr11_+_68208554
|
0.669
|
NM_015973
|
GAL
|
galanin prepropeptide
|
|
chr19_-_54618410
|
0.669
|
NM_178449
|
PTH2
|
parathyroid hormone 2
|
|
chr1_-_52116598
|
0.668
|
|
NRD1
|
nardilysin (N-arginine dibasic convertase)
|
|
chr6_-_31779082
|
0.667
|
|
ABHD16A
|
abhydrolase domain containing 16A
|
|
chr7_+_116447527
|
0.665
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr11_-_6589976
|
0.663
|
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chr1_+_110329224
|
0.661
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr1_-_227710619
|
0.659
|
|
NUP133
|
nucleoporin 133kDa
|
|
chr1_+_51474502
|
0.657
|
|
RNF11
|
ring finger protein 11
|
|
chr16_+_2742653
|
0.657
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr8_+_42515873
|
0.656
|
NM_138436
NM_001135674
NM_001135675
|
C8orf40
|
chromosome 8 open reading frame 40
|
|
chr1_+_177529559
|
0.655
|
|
SOAT1
|
sterol O-acyltransferase 1
|
|
chr11_+_68208583
|
0.654
|
|
GAL
|
galanin prepropeptide
|
|
chr19_-_50779914
|
0.651
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr6_-_31779099
|
0.651
|
NM_021160
|
ABHD16A
|
abhydrolase domain containing 16A
|
|
chr1_+_36462625
|
0.650
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr2_-_87906400
|
0.646
|
NM_001078170
|
RGPD2
|
RANBP2-like and GRIP domain containing 2
|
|
chr7_-_158314945
|
0.642
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr1_-_201194033
|
0.642
|
|
ADIPOR1
|
adiponectin receptor 1
|
|
chr1_-_184611012
|
0.641
|
NM_003292
|
TPR
|
translocated promoter region (to activated MET oncogene)
|
|
chrX_-_54087331
|
0.640
|
NM_001184897
NM_015107
|
PHF8
|
PHD finger protein 8
|
|
chrX_+_151991474
|
0.636
|
NM_001170944
NM_032882
|
LOC100287428
PNMA6A
|
paraneoplastic antigen-like protein 6A related gene
paraneoplastic antigen like 6A
|
|
chr1_+_177529511
|
0.636
|
|
SOAT1
|
sterol O-acyltransferase 1
|
|
chr3_-_10042989
|
0.633
|
|
CIDECP
|
cell death-inducing DFFA-like effector c pseudogene
|
|
chr8_+_26204997
|
0.631
|
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha
|
|
chr6_+_36670113
|
0.630
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr11_-_116474196
|
0.628
|
NM_025164
|
SIK3
|
SIK family kinase 3
|
|
chr6_-_31778825
|
0.628
|
NM_001177515
|
ABHD16A
|
abhydrolase domain containing 16A
|
|
chr2_-_220116508
|
0.621
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr3_+_45705681
|
0.620
|
NM_014016
|
SACM1L
|
SAC1 suppressor of actin mutations 1-like (yeast)
|
|
chr3_-_49041862
|
0.619
|
NM_000884
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr3_+_10003579
|
0.617
|
|
LOC442075
|
hypothetical LOC442075
|
|
chr3_-_49041817
|
0.617
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr7_+_89813913
|
0.616
|
NM_001042717
NM_033107
|
GTPBP10
|
GTP-binding protein 10 (putative)
|
|
chr4_-_8493327
|
0.616
|
NM_001101667
NM_003501
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr1_+_51474531
|
0.616
|
NM_014372
|
RNF11
|
ring finger protein 11
|
|
chr12_-_74764614
|
0.615
|
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr1_-_227710700
|
0.613
|
NM_018230
|
NUP133
|
nucleoporin 133kDa
|
|
chr7_+_38184363
|
0.608
|
|
STARD3NL
|
STARD3 N-terminal like
|
|
chr11_-_6590004
|
0.607
|
NM_006284
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chr1_+_40930080
|
0.606
|
|
NFYC
|
nuclear transcription factor Y, gamma
|
|
chr7_+_107318821
|
0.605
|
NM_000108
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr6_+_36670117
|
0.605
|
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr5_+_78567604
|
0.604
|
NM_152405
|
JMY
|
junction mediating and regulatory protein, p53 cofactor
|
|
chr7_+_89813969
|
0.602
|
|
GTPBP10
|
GTP-binding protein 10 (putative)
|
|
chr5_-_54639158
|
0.601
|
NM_019030
|
DHX29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29
|
|
chrX_-_15263586
|
0.600
|
|
PIGA
|
phosphatidylinositol glycan anchor biosynthesis, class A
|
|
chr3_-_25806500
|
0.599
|
NM_001145294
|
NGLY1
|
N-glycanase 1
|
|
chr6_+_36670067
|
0.599
|
NM_003017
|
SRSF3
|
serine/arginine-rich splicing factor 3
|
|
chr6_-_29703725
|
0.596
|
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr7_-_106991941
|
0.595
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr22_-_18484744
|
0.593
|
NM_022727
NM_182984
|
TRMT2A
|
TRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae)
|
|
chr10_+_21863272
|
0.592
|
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr15_-_29950188
|
0.592
|
|
OTUD7A
|
OTU domain containing 7A
|
|
chr7_+_107318854
|
0.591
|
|
DLD
|
dihydrolipoamide dehydrogenase
|
|
chr5_+_68425672
|
0.591
|
|
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5
|
|
chr1_-_226712140
|
0.590
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|