|
chr1_+_190811479
|
1.974
|
NM_002922
|
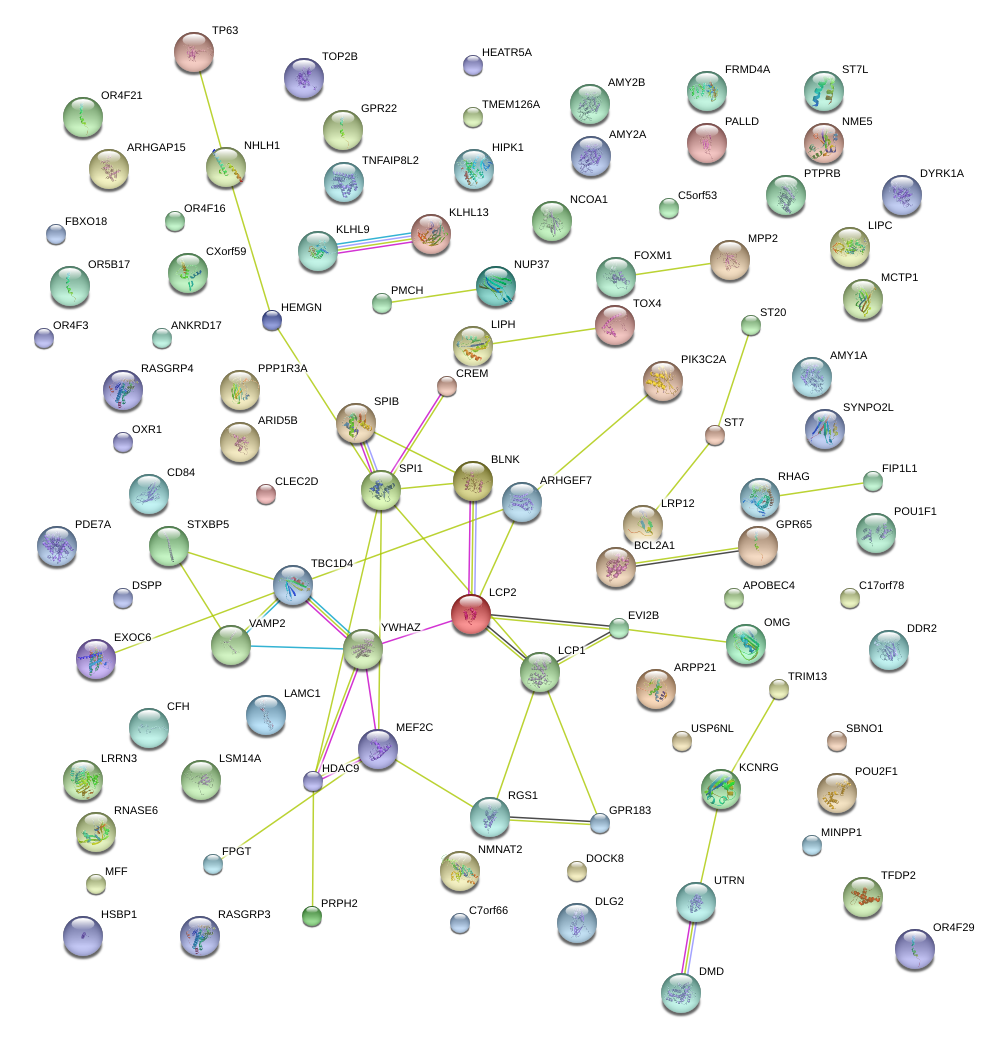
RGS1
|
regulator of G-protein signaling 1
|
|
chrX_-_33139349
|
1.871
|
NM_004006
|
DMD
|
dystrophin
|
|
chrX_-_117003310
|
1.698
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr5_-_88155360
|
1.624
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_+_24660849
|
1.347
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr3_-_87408301
|
1.237
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr12_-_101115677
|
1.157
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr10_-_98021144
|
1.115
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr2_+_143603368
|
1.103
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr8_-_102005697
|
1.055
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr11_-_17147863
|
1.034
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr12_+_9713472
|
1.012
|
NM_001197318
NM_013269
NM_001004419
NM_001197317
NM_001197319
|
CLEC2D
|
C-type lectin domain family 2, member D
|
|
chr13_+_49487390
|
1.004
|
NM_173605
NM_199464
|
TRIM13
KCNRG
|
tripartite motif containing 13
potassium channel regulator
|
|
chr14_+_20319049
|
0.997
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr1_+_165565006
|
0.957
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr7_+_110518297
|
0.943
|
NM_001099658
NM_001099660
NM_018334
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr4_+_169654742
|
0.941
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr8_+_107807415
|
0.926
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr5_+_76470829
|
0.924
|
|
|
|
|
chr1_+_194887630
|
0.914
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr10_+_5976354
|
0.904
|
NM_032807
|
FBXO18
|
F-box protein, helicase, 18
|
|
chr5_+_180726874
|
0.899
|
NM_001005224
NM_001005221
NM_001005277
|
OR4F3
OR4F29
OR4F16
|
olfactory receptor, family 4, subfamily F, member 3
olfactory receptor, family 4, subfamily F, member 29
olfactory receptor, family 4, subfamily F, member 16
|
|
chr10_-_14090459
|
0.873
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr4_+_88748704
|
0.870
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr20_-_42561725
|
0.864
|
|
|
|
|
chr1_+_74473672
|
0.856
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr15_-_78050515
|
0.849
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr10_+_89257569
|
0.832
|
NM_001178118
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr3_+_190831909
|
0.809
|
NM_001114978
NM_001114979
NM_003722
|
TP63
|
tumor protein p63
|
|
chrX_+_35974973
|
0.803
|
NM_173695
|
CXorf59
|
chromosome X open reading frame 59
|
|
chr19_+_39402167
|
0.803
|
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae)
|
|
chr7_+_116447527
|
0.802
|
|
ST7
|
suppression of tumorigenicity 7
|
|
chr1_-_181540631
|
0.802
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr17_-_26648375
|
0.800
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr9_-_99740243
|
0.798
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr10_+_35524835
|
0.797
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr2_+_143603439
|
0.796
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr2_+_227900471
|
0.793
|
NM_020194
|
MFF
|
mitochondrial fission factor
|
|
chr5_-_169657192
|
0.777
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr5_-_169657395
|
0.777
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr1_+_158603480
|
0.775
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr14_+_87541220
|
0.769
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr10_+_94584449
|
0.766
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr5_+_139485703
|
0.765
|
NM_001007189
|
C5orf53
|
chromosome 5 open reading frame 53
|
|
chr7_+_18502342
|
0.757
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr5_-_169657312
|
0.755
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr4_-_74307637
|
0.753
|
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr10_-_75080784
|
0.744
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr12_-_101036407
|
0.740
|
NM_024057
|
NUP37
|
nucleoporin 37kDa
|
|
chr3_-_42818221
|
0.737
|
|
|
|
|
chr8_-_66863841
|
0.736
|
NM_002603
|
PDE7A
|
phosphodiesterase 7A
|
|
chr11_-_47356479
|
0.729
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr10_-_11614279
|
0.724
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr1_+_114295289
|
0.720
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr1_+_160868932
|
0.714
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr19_+_55614006
|
0.705
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr9_+_263025
|
0.695
|
NM_001190458
NM_001193536
|
DOCK8
|
dedicator of cytokinesis 8
|
|
chr1_+_149395728
|
0.694
|
NM_024575
|
TNFAIP8L2
|
tumor necrosis factor, alpha-induced protein 8-like 2
|
|
chr5_-_94442941
|
0.694
|
|
MCTP1
|
multiple C2 domains, transmembrane 1
|
|
chr19_-_43608641
|
0.682
|
NM_001146202
NM_001146203
NM_001146204
NM_001146205
NM_001146206
NM_001146207
NM_170604
|
RASGRP4
|
RAS guanyl releasing protein 4
|
|
chr7_-_113346284
|
0.682
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A
|
|
chr3_-_143230124
|
0.681
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr14_-_30927931
|
0.671
|
NM_015473
|
HEATR5A
|
HEAT repeat containing 5A
|
|
chr17_+_32807040
|
0.648
|
NM_173625
|
C17orf78
|
chromosome 17 open reading frame 78
|
|
chr11_-_47356650
|
0.640
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr3_-_186752952
|
0.637
|
NM_139248
|
LIPH
|
lipase, member H
|
|
chr12_-_69317441
|
0.630
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr5_-_169657360
|
0.629
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr12_-_122400937
|
0.627
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr13_-_98757634
|
0.616
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr10_+_63331018
|
0.615
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr4_+_53975597
|
0.611
|
|
FIP1L1
|
FIP1 like 1 (S. cerevisiae)
|
|
chr1_-_181889070
|
0.610
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr1_+_181380837
|
0.603
|
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr11_-_47356642
|
0.603
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr1_+_160868822
|
0.599
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr11_-_57883117
|
0.599
|
NM_001005489
|
OR5B17
|
olfactory receptor, family 5, subfamily B, member 17
|
|
chr12_+_63510481
|
0.590
|
|
|
|
|
chr11_-_47356672
|
0.590
|
NM_001080547
NM_003120
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr11_-_47356562
|
0.582
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr5_-_137502975
|
0.579
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr7_+_106897737
|
0.574
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr1_-_158815895
|
0.574
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr6_-_49712483
|
0.565
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr2_+_33592445
|
0.564
|
NM_015376
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr17_-_8004671
|
0.562
|
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr1_+_103898844
|
0.561
|
NM_020978
|
AMY2B
|
amylase, alpha 2B (pancreatic)
|
|
chr13_+_110653407
|
0.556
|
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr21_+_37713076
|
0.551
|
NM_130436
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr13_-_74799898
|
0.549
|
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr13_-_45640679
|
0.548
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr6_+_147568795
|
0.546
|
|
STXBP5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr3_-_25658829
|
0.545
|
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa
|
|
chr3_+_35658852
|
0.545
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr7_-_108311872
|
0.544
|
NM_001024607
|
C7orf66
|
chromosome 7 open reading frame 66
|
|
chr17_-_26665220
|
0.543
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr11_-_83071084
|
0.542
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr7_+_142723340
|
0.541
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr3_+_44665220
|
0.540
|
NM_003420
|
ZNF35
|
zinc finger protein 35
|
|
chr17_-_26665213
|
0.539
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr4_-_38710376
|
0.538
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr5_+_139869819
|
0.536
|
|
|
|
|
chr1_-_180822686
|
0.534
|
NM_021133
|
RNASEL
|
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent)
|
|
chr6_-_133120587
|
0.529
|
NM_004665
|
VNN2
|
vanin 2
|
|
chr15_+_38296920
|
0.527
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr15_-_23204887
|
0.523
|
NM_000462
|
UBE3A
|
ubiquitin protein ligase E3A
|
|
chr1_+_84540921
|
0.523
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr5_+_67622242
|
0.522
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr21_-_14840506
|
0.520
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr8_+_67568044
|
0.517
|
NM_152765
|
C8orf46
|
chromosome 8 open reading frame 46
|
|
chr13_-_40454360
|
0.516
|
NM_001145353
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr3_-_152585226
|
0.515
|
NM_022788
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chr1_-_178100204
|
0.510
|
|
|
|
|
chr2_+_151922350
|
0.508
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr1_+_62835759
|
0.508
|
NM_014495
|
ANGPTL3
|
angiopoietin-like 3
|
|
chr12_+_21417068
|
0.507
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr2_-_55053398
|
0.507
|
|
|
|
|
chrX_-_131989963
|
0.505
|
NM_031907
|
USP26
|
ubiquitin specific peptidase 26
|
|
chr1_+_242281199
|
0.505
|
|
ZNF238
|
zinc finger protein 238
|
|
chr14_-_94669547
|
0.498
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr1_+_40635058
|
0.494
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chrX_-_138742050
|
0.492
|
NM_001010986
NM_173694
|
ATP11C
|
ATPase, class VI, type 11C
|
|
chr12_+_25096472
|
0.490
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr7_+_129634935
|
0.490
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr13_-_98708549
|
0.489
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr2_-_207291364
|
0.487
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr17_-_26665194
|
0.480
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr1_+_151015471
|
0.479
|
NM_178354
|
LCE1F
|
late cornified envelope 1F
|
|
chr7_+_113853790
|
0.478
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr9_-_21472311
|
0.478
|
NM_176891
|
IFNE
|
interferon, epsilon
|
|
chr11_-_5321329
|
0.476
|
NM_001005567
|
OR51B5
|
olfactory receptor, family 51, subfamily B, member 5
|
|
chr10_-_61570665
|
0.475
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr14_-_80475636
|
0.474
|
NM_152446
|
C14orf145
|
chromosome 14 open reading frame 145
|
|
chr11_-_72057755
|
0.473
|
NM_001146209
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr3_+_44667504
|
0.473
|
|
ZNF35
|
zinc finger protein 35
|
|
chr1_+_196874759
|
0.471
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr8_-_102872372
|
0.471
|
|
NCALD
|
neurocalcin delta
|
|
chr14_-_101617680
|
0.470
|
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1
|
|
chr1_-_239865724
|
0.465
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chr5_-_43326922
|
0.463
|
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble)
|
|
chr2_+_161701711
|
0.463
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr3_-_195670256
|
0.463
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr17_-_36209672
|
0.461
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr4_+_73123712
|
0.459
|
NM_001144756
NM_053036
|
NPFFR2
|
neuropeptide FF receptor 2
|
|
chr10_-_97190885
|
0.458
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr2_-_55090759
|
0.458
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr1_+_78243181
|
0.455
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr17_-_26665144
|
0.455
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr17_-_1195096
|
0.455
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr10_+_50177192
|
0.455
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr4_+_71492581
|
0.448
|
NM_016519
|
AMBN
|
ameloblastin (enamel matrix protein)
|
|
chr1_-_246869181
|
0.447
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr2_+_211129567
|
0.445
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr6_-_32892705
|
0.442
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr8_-_102872614
|
0.441
|
NM_032041
|
NCALD
|
neurocalcin delta
|
|
chr21_-_30836051
|
0.437
|
NM_181612
|
KRTAP19-6
|
keratin associated protein 19-6
|
|
chr3_-_17757402
|
0.437
|
NM_014744
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr9_+_19280748
|
0.437
|
NM_017925
|
DENND4C
|
DENN/MADD domain containing 4C
|
|
chr2_-_55413253
|
0.436
|
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr6_+_135558744
|
0.434
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr8_-_102872354
|
0.432
|
|
NCALD
|
neurocalcin delta
|
|
chr4_+_77214886
|
0.430
|
NM_001130016
NM_001179
|
ART3
|
ADP-ribosyltransferase 3
|
|
chr4_+_71713324
|
0.429
|
NM_031889
|
ENAM
|
enamelin
|
|
chr4_+_71283383
|
0.425
|
NM_006685
|
SMR3B
|
submaxillary gland androgen regulated protein 3B
|
|
chr7_-_135263737
|
0.425
|
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chr11_+_33319680
|
0.424
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr5_+_156540425
|
0.424
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chrX_+_65300803
|
0.421
|
NM_001130860
NM_014799
|
HEPH
|
hephaestin
|
|
chr13_+_45174446
|
0.420
|
NM_152719
|
SPERT
|
spermatid associated
|
|
chr1_+_84539876
|
0.418
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr15_-_75135496
|
0.417
|
|
TSPAN3
|
tetraspanin 3
|
|
chr1_+_207926172
|
0.416
|
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr17_+_58440629
|
0.416
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr4_-_146315341
|
0.415
|
NM_017493
|
OTUD4
|
OTU domain containing 4
|
|
chr17_+_23824784
|
0.415
|
NM_001145975
NM_001145976
NM_003984
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr2_-_162717159
|
0.414
|
NM_002054
|
GCG
|
glucagon
|
|
chr2_+_33514896
|
0.412
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr6_-_32665409
|
0.411
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr18_-_65766068
|
0.411
|
|
CD226
|
CD226 molecule
|
|
chrX_-_135691168
|
0.409
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr1_+_157167965
|
0.408
|
NM_152501
NM_198928
NM_198929
NM_198930
|
PYHIN1
|
pyrin and HIN domain family, member 1
|
|
chr17_-_36276987
|
0.403
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr1_-_150398327
|
0.402
|
NM_001122965
|
RPTN
|
repetin
|
|
chr15_-_42756377
|
0.401
|
NM_001145112
|
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast)
|
|
chr5_-_58918031
|
0.401
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_52281050
|
0.400
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr3_+_113534604
|
0.400
|
NM_001004196
NM_005944
|
CD200
|
CD200 molecule
|
|
chr10_-_21226536
|
0.400
|
NM_006393
|
NEBL
|
nebulette
|
|
chr16_-_54466709
|
0.399
|
NM_001143685
NM_145024
|
CES5A
|
carboxylesterase 5A
|
|
chr21_+_37714470
|
0.394
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr10_+_98054074
|
0.393
|
NM_001017520
NM_004088
|
DNTT
|
deoxynucleotidyltransferase, terminal
|
|
chr3_-_147696411
|
0.393
|
NM_020359
|
PLSCR2
|
phospholipid scramblase 2
|
|
chr2_-_37150876
|
0.393
|
|
HEATR5B
|
HEAT repeat containing 5B
|
|
chr21_-_31995972
|
0.392
|
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr6_-_117853710
|
0.392
|
NM_002944
|
ROS1
|
c-ros oncogene 1 , receptor tyrosine kinase
|
|
chrX_+_41468351
|
0.389
|
NM_080817
|
GPR82
|
G protein-coupled receptor 82
|
|
chr21_-_36870736
|
0.386
|
NM_001146077
|
CLDN14
|
claudin 14
|
|
chr22_+_27779877
|
0.383
|
|
ZNRF3
|
zinc and ring finger 3
|
|
chr10_-_31328426
|
0.382
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr12_+_130004404
|
0.377
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|