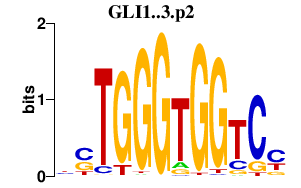
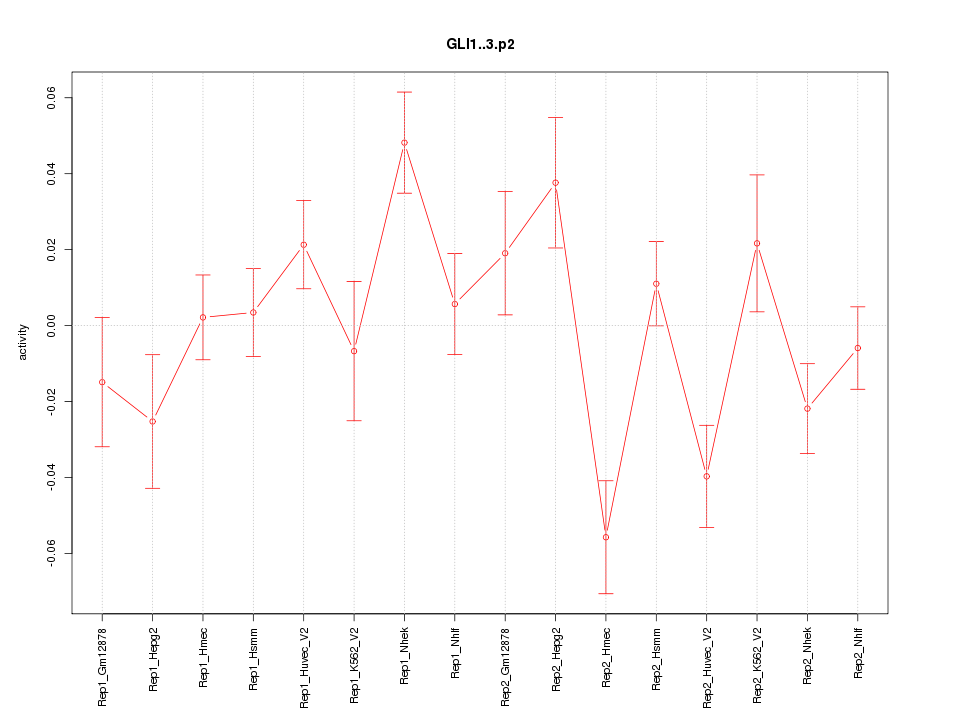
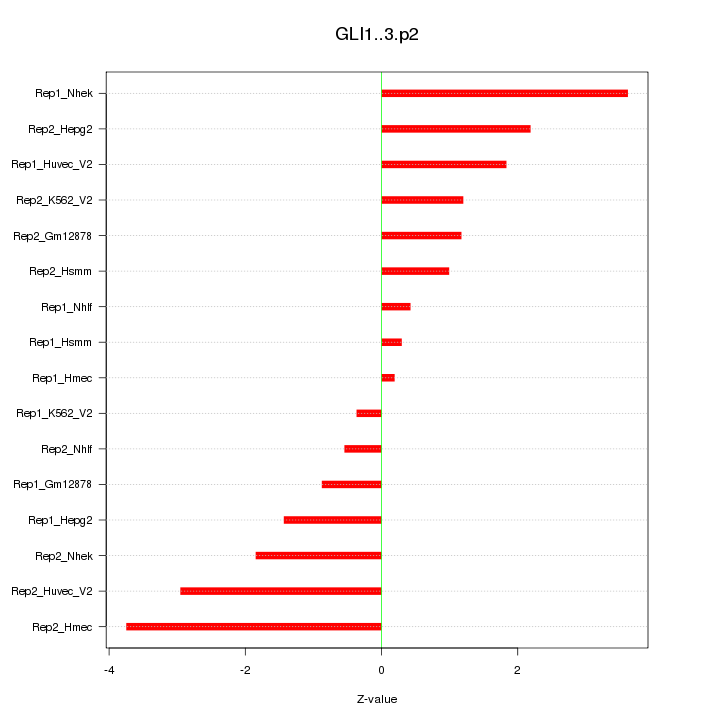
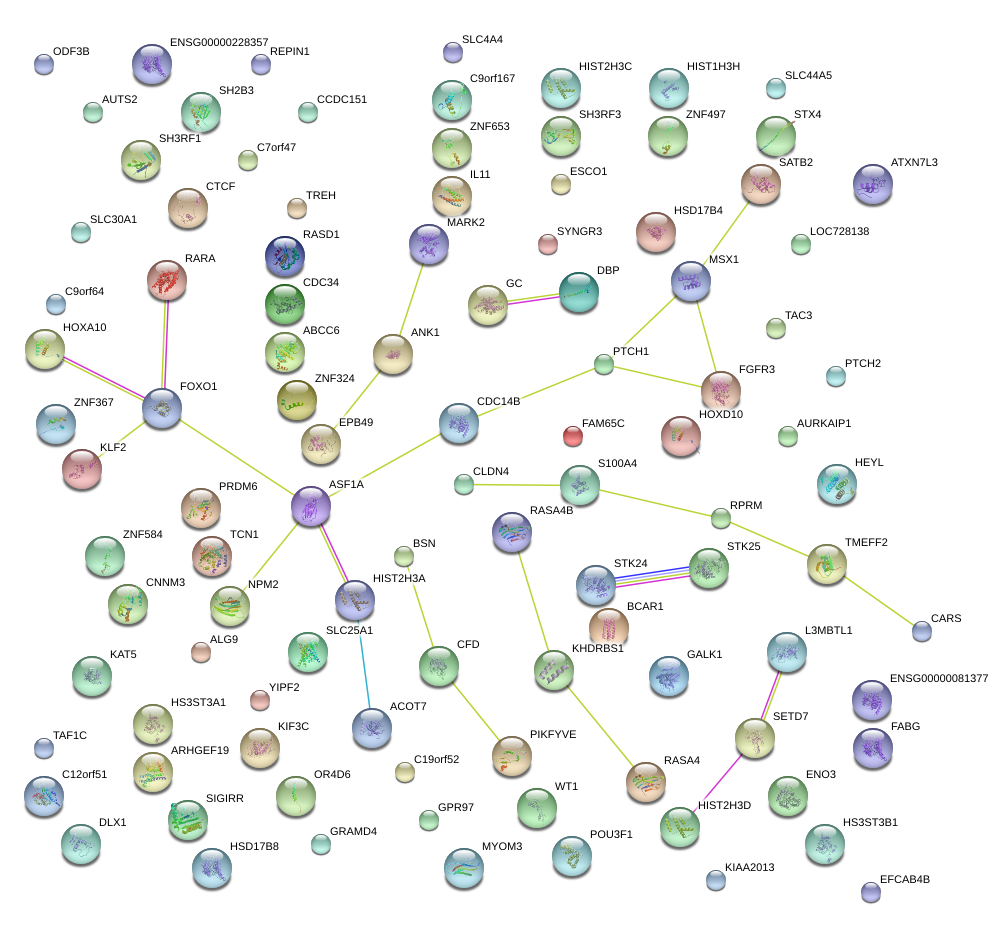
|
chr7_+_149699185
|
1.245
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr1_-_75854281
|
1.118
|
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr12_-_3732614
|
1.077
|
NM_001144958
NM_001144959
NM_032680
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr16_-_3049336
|
1.076
|
|
|
|
|
chr19_-_10900086
|
1.048
|
NM_024029
|
YIPF2
|
Yip1 domain family, member 2
|
|
chr20_-_48686832
|
0.986
|
NM_080829
|
FAM65C
|
family with sequence similarity 65, member C
|
|
chr1_-_11909042
|
0.974
|
NM_138346
|
KIAA2013
|
KIAA2013
|
|
chr4_+_1764802
|
0.964
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr16_+_66153810
|
0.961
|
NM_001191022
NM_006565
|
CTCF
|
CCCTC-binding factor (zinc finger protein)
|
|
chr14_+_104218487
|
0.941
|
|
|
|
|
chr19_+_10900419
|
0.937
|
NM_138358
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr2_-_192767844
|
0.933
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr1_-_209818675
|
0.920
|
NM_021194
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr11_+_58980986
|
0.909
|
NM_001004708
|
OR4D6
|
olfactory receptor, family 4, subfamily D, member 6
|
|
chr12_-_3732478
|
0.890
|
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr6_+_44076281
|
0.879
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr1_-_38285033
|
0.879
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr1_+_148090804
|
0.877
|
NM_021059
NM_001005464
|
HIST2H3C
HIST2H3A
|
histone cluster 2, H3c
histone cluster 2, H3a
|
|
chr4_+_72271218
|
0.866
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr11_-_404975
|
0.849
|
NM_001135054
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr19_+_10900502
|
0.833
|
|
C19orf52
|
chromosome 19 open reading frame 52
|
|
chr11_-_118055564
|
0.827
|
NM_007180
|
TREH
|
trehalase (brush-border membrane glycoprotein)
|
|
chr13_-_40138607
|
0.802
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr1_-_209818576
|
0.789
|
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr19_+_38485583
|
0.778
|
|
LOC80054
|
hypothetical LOC80054
|
|
chr7_+_68702252
|
0.773
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr16_-_82778071
|
0.772
|
NM_005679
NM_139353
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa
|
|
chr22_-_49317408
|
0.759
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr2_-_154043403
|
0.759
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr3_+_49566901
|
0.757
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr2_-_26058831
|
0.753
|
|
KIF3C
|
kinesin family member 3C
|
|
chr11_-_3035172
|
0.748
|
NM_001014437
NM_001194997
NM_001751
NM_139273
|
CARS
|
cysteinyl-tRNA synthetase
|
|
chr1_-_151784889
|
0.746
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr1_-_16411690
|
0.737
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr1_-_45081190
|
0.732
|
NM_001166292
NM_003738
|
PTCH2
|
patched 2
|
|
chr1_-_6343306
|
0.722
|
NM_181865
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr16_+_1979959
|
0.698
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr7_+_72883128
|
0.697
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr19_-_63565890
|
0.691
|
NM_198458
|
ZNF497
|
zinc finger protein 497
|
|
chr4_+_4912272
|
0.680
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr2_-_242096673
|
0.677
|
NM_006374
|
STK25
|
serine/threonine kinase 25
|
|
chr9_-_97318760
|
0.668
|
|
PTCH1
|
patched 1
|
|
chr8_+_21972804
|
0.668
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr11_+_9736408
|
0.660
|
|
LOC283104
|
hypothetical LOC283104
|
|
chr17_-_39632845
|
0.659
|
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr17_+_35751796
|
0.652
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr6_+_33280396
|
0.651
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chr4_-_140696740
|
0.643
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr1_-_39877928
|
0.642
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr9_+_8848130
|
0.639
|
|
|
|
|
chr19_+_16296632
|
0.631
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr7_-_23476156
|
0.629
|
|
|
|
|
chr17_-_17340219
|
0.625
|
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr16_+_56259657
|
0.625
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr1_-_1300680
|
0.624
|
NM_017900
|
AURKAIP1
|
aurora kinase A interacting protein 1
|
|
chr8_+_21937533
|
0.621
|
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr17_+_71141108
|
0.618
|
NM_001162995
|
LOC643008
|
hypothetical protein LOC643008
|
|
chr12_+_110328126
|
0.613
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr1_-_39877883
|
0.606
|
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|
|
chr22_+_46406115
|
0.603
|
|
LOC284930
|
hypothetical protein LOC284930
|
|
chr8_+_21972631
|
0.602
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr9_+_139292094
|
0.602
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr1_-_24307514
|
0.601
|
|
MYOM3
|
myomesin family, member 3
|
|
chr7_-_99871981
|
0.599
|
NM_145030
|
C7orf47
|
chromosome 7 open reading frame 47
|
|
chr19_-_11477689
|
0.598
|
NM_138783
|
ZNF653
|
zinc finger protein 653
|
|
chr2_+_109112428
|
0.596
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr5_+_122452739
|
0.595
|
NM_001136239
|
PRDM6
|
PR domain containing 6
|
|
chr20_+_41576459
|
0.584
|
NM_015478
|
L3MBTL1
|
l(3)mbt-like 1 (Drosophila)
|
|
chr19_-_60573552
|
0.582
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr2_+_172658453
|
0.580
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr11_-_32413486
|
0.580
|
|
WT1
|
Wilms tumor 1
|
|
chr16_+_30952350
|
0.578
|
|
STX4
|
syntaxin 4
|
|
chr14_+_106009489
|
0.577
|
|
NCRNA00221
|
non-protein coding RNA 221
|
|
chr6_+_19946591
|
0.575
|
|
|
|
|
chr22_+_18090118
|
0.572
|
|
SEPT5-GP1BB
|
SEPT5-GP1BB read-through transcript
|
|
chr19_+_16296734
|
0.570
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr11_+_65236261
|
0.567
|
|
KAT5
|
K(lysine) acetyltransferase 5
|
|
chr17_+_4795128
|
0.565
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|
|
chr22_+_45401321
|
0.563
|
NM_015124
|
GRAMD4
|
GRAM domain containing 4
|
|
chr12_-_111304138
|
0.562
|
NM_001109662
|
C12orf51
|
chromosome 12 open reading frame 51
|
|
chr7_+_68701704
|
0.557
|
NM_001127231
NM_001127232
NM_015570
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr7_-_102044374
|
0.556
|
|
RASA4
|
RAS p21 protein activator 4
|
|
chr17_-_60401967
|
0.555
|
|
AMZ2P1
|
archaelysin family metallopeptidase 2 pseudogene 1
|
|
chr19_+_63670148
|
0.553
|
NM_014347
|
ZNF324
|
zinc finger protein 324
|
|
chr17_-_71272811
|
0.553
|
NM_000154
|
GALK1
|
galactokinase 1
|
|
chr18_-_17434811
|
0.552
|
|
ESCO1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae)
|
|
chr16_-_73842884
|
0.547
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr2_-_200030944
|
0.545
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr1_+_22224571
|
0.543
|
|
HSPC157
|
hypothetical LOC29092
|
|
chr1_+_32251988
|
0.543
|
NM_006559
|
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1
|
|
chr7_-_27180398
|
0.539
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr2_+_96845704
|
0.536
|
NM_017623
NM_199078
|
CNNM3
|
cyclin M3
|
|
chr12_-_55696542
|
0.535
|
NM_001178054
NM_013251
|
TAC3
|
tachykinin 3
|
|
chr17_+_14145051
|
0.533
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr16_-_16224791
|
0.531
|
NM_001079528
NM_001171
|
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6
|
|
chr19_+_2240996
|
0.530
|
|
|
|
|
chr8_-_41774296
|
0.529
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr22_-_17545872
|
0.529
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr9_-_85761472
|
0.527
|
NM_032307
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr19_+_811615
|
0.522
|
|
CFD
|
complement factor D (adipsin)
|
|
chr19_+_482737
|
0.522
|
|
CDC34
|
cell division cycle 34 homolog (S. cerevisiae)
|
|
chr19_-_53832381
|
0.521
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr9_-_98368913
|
0.519
|
NM_001077181
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr2_+_19932101
|
0.517
|
|
FLJ12334
|
hypothetical LOC400946
|
|
chr6_+_3009120
|
0.516
|
|
RIPK1
|
receptor (TNFRSF)-interacting serine-threonine kinase 1
|
|
chr12_-_55316350
|
0.514
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr18_+_13208777
|
0.511
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr16_+_30952382
|
0.510
|
NM_004604
|
STX4
|
syntaxin 4
|
|
chr15_+_38520594
|
0.509
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr2_+_220200530
|
0.508
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chr17_-_8474733
|
0.507
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr5_-_177950161
|
0.506
|
NM_173465
|
COL23A1
|
collagen, type XXIII, alpha 1
|
|
chr14_-_73296387
|
0.504
|
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr17_+_45008473
|
0.503
|
|
NXPH3
|
neurexophilin 3
|
|
chr16_+_67778441
|
0.501
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr16_-_2521359
|
0.497
|
NM_001048212
|
CEMP1
|
cementum protein 1
|
|
chr16_+_705080
|
0.494
|
NM_024042
|
METRN
|
meteorin, glial cell differentiation regulator
|
|
chr7_-_1034453
|
0.493
|
|
C7orf50
|
chromosome 7 open reading frame 50
|
|
chr2_-_242096592
|
0.486
|
|
STK25
|
serine/threonine kinase 25
|
|
chr7_-_102044419
|
0.486
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chr19_-_54618410
|
0.485
|
NM_178449
|
PTH2
|
parathyroid hormone 2
|
|
chr21_-_32906729
|
0.484
|
NM_021254
|
C21orf59
|
chromosome 21 open reading frame 59
|
|
chr11_-_18913121
|
0.481
|
NM_147199
|
MRGPRX1
|
MAS-related GPR, member X1
|
|
chr14_-_103058568
|
0.480
|
|
CKB
|
creatine kinase, brain
|
|
chr2_+_178685396
|
0.478
|
NM_152945
|
RBM45
|
RNA binding motif protein 45
|
|
chr17_-_37181626
|
0.478
|
|
|
|
|
chr16_+_85169615
|
0.476
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr18_-_42798364
|
0.474
|
NM_001100817
|
TCEB3C
TCEB3CL
|
transcription elongation factor B polypeptide 3C (elongin A3)
transcription elongation factor B polypeptide 3C-like
|
|
chr17_+_37693090
|
0.473
|
NM_003152
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr8_+_31616809
|
0.473
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr1_+_11646735
|
0.472
|
NM_018438
|
FBXO6
|
F-box protein 6
|
|
chr22_-_17546276
|
0.472
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr11_-_65871592
|
0.471
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr22_+_18388656
|
0.470
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr22_-_17546234
|
0.469
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr16_-_2258067
|
0.467
|
|
RNPS1
|
RNA binding protein S1, serine-rich domain
|
|
chr17_-_34811377
|
0.466
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr10_+_52420936
|
0.464
|
NM_001098512
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr17_-_1342700
|
0.464
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr8_-_10550026
|
0.463
|
NM_178857
|
RP1L1
|
retinitis pigmentosa 1-like 1
|
|
chr5_-_179712920
|
0.463
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr1_-_152202574
|
0.463
|
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1
|
|
chr20_-_48741481
|
0.461
|
|
|
|
|
chr3_+_50272648
|
0.460
|
|
|
|
|
chr19_+_63654766
|
0.459
|
NM_207395
|
ZNF324B
|
zinc finger protein 324B
|
|
chr17_+_7283412
|
0.457
|
NM_004112
|
FGF11
|
fibroblast growth factor 11
|
|
chr22_-_17546296
|
0.456
|
NM_005984
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr2_+_96355661
|
0.455
|
NM_178495
NM_001163524
|
ITPRIPL1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1
|
|
chr20_-_34102250
|
0.455
|
|
LOC647979
|
hypothetical LOC647979
|
|
chr2_-_6923215
|
0.454
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr5_+_176449156
|
0.453
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr16_+_29581783
|
0.452
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr9_-_85761451
|
0.452
|
|
C9orf64
|
chromosome 9 open reading frame 64
|
|
chr17_+_45703765
|
0.451
|
NM_001168215
|
TMEM92
|
transmembrane protein 92
|
|
chr7_-_126819953
|
0.450
|
NM_176814
|
ZNF800
|
zinc finger protein 800
|
|
chr14_+_76297885
|
0.445
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr22_-_49050847
|
0.445
|
NM_002751
|
MAPK11
|
mitogen-activated protein kinase 11
|
|
chr1_-_16812274
|
0.445
|
NM_017940
|
NBPF1
|
neuroblastoma breakpoint family, member 1
|
|
chr22_+_18388633
|
0.444
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr17_-_7238546
|
0.444
|
NM_020360
|
PLSCR3
|
phospholipid scramblase 3
|
|
chr7_-_23476497
|
0.444
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr5_+_71438796
|
0.442
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr8_-_142080169
|
0.440
|
NM_005607
NM_153831
|
PTK2
|
PTK2 protein tyrosine kinase 2
|
|
chr1_-_32574185
|
0.439
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr10_+_52421120
|
0.438
|
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr11_+_122806196
|
0.438
|
|
LOC100128242
|
hypothetical LOC100128242
|
|
chr11_+_10434056
|
0.438
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr19_-_48700824
|
0.438
|
NM_198850
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3
|
|
chr2_+_149119029
|
0.434
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr1_-_39929654
|
0.433
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr5_-_1397941
|
0.432
|
NM_030782
|
CLPTM1L
|
CLPTM1-like
|
|
chr14_+_103165244
|
0.432
|
NM_001130107
NM_005552
NM_182923
|
KLC1
|
kinesin light chain 1
|
|
chr2_+_173927783
|
0.431
|
NM_031942
NM_145810
|
CDCA7
|
cell division cycle associated 7
|
|
chr10_+_129237602
|
0.430
|
NM_001030013
|
NPS
|
neuropeptide S
|
|
chr7_+_119700924
|
0.429
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr19_+_935316
|
0.429
|
NM_024100
|
WDR18
|
WD repeat domain 18
|
|
chr16_-_29818334
|
0.425
|
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr1_-_1300385
|
0.425
|
|
AURKAIP1
|
aurora kinase A interacting protein 1
|
|
chr3_+_3816079
|
0.424
|
NM_020873
|
LRRN1
|
leucine rich repeat neuronal 1
|
|
chr15_-_38834740
|
0.424
|
NM_018145
|
FAM82A2
|
family with sequence similarity 82, member A2
|
|
chr11_-_64268126
|
0.423
|
NM_001098671
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr20_+_61655934
|
0.420
|
|
|
|
|
chr1_-_1300416
|
0.417
|
NM_001127230
NM_001127229
|
AURKAIP1
|
aurora kinase A interacting protein 1
|
|
chr6_-_3697244
|
0.417
|
NM_183373
|
C6orf145
|
chromosome 6 open reading frame 145
|
|
chr12_-_6680053
|
0.417
|
NM_153685
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
chr17_-_3740785
|
0.416
|
NM_172206
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha
|
|
chr1_+_26986996
|
0.416
|
NM_017837
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V
|
|
chr19_+_60851207
|
0.415
|
|
CCDC106
|
coiled-coil domain containing 106
|
|
chr16_+_48865521
|
0.415
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr14_+_99181232
|
0.414
|
NM_001127258
NM_032425
|
HHIPL1
|
HHIP-like 1
|
|
chr9_+_130620540
|
0.414
|
NM_004435
|
ENDOG
|
endonuclease G
|
|
chr11_+_63766163
|
0.413
|
NM_057092
|
FKBP2
|
FK506 binding protein 2, 13kDa
|
|
chr20_+_31783426
|
0.411
|
NM_032819
|
ZNF341
|
zinc finger protein 341
|
|
chr5_+_175725050
|
0.408
|
NM_173664
|
ARL10
|
ADP-ribosylation factor-like 10
|
|
chr1_+_28568643
|
0.408
|
NM_001048183
|
PHACTR4
|
phosphatase and actin regulator 4
|
|
chr11_+_76455574
|
0.408
|
NM_004055
|
CAPN5
|
calpain 5
|
|
chr3_-_193609531
|
0.407
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr20_-_36095236
|
0.407
|
NM_014657
|
TTI1
|
Tel2 interacting protein 1 homolog (S. pombe)
|
|
chr3_-_50580066
|
0.406
|
NM_001171740
NM_001171743
NM_016210
|
C3orf18
|
chromosome 3 open reading frame 18
|
|
chr4_-_5071996
|
0.406
|
NM_018659
|
CYTL1
|
cytokine-like 1
|