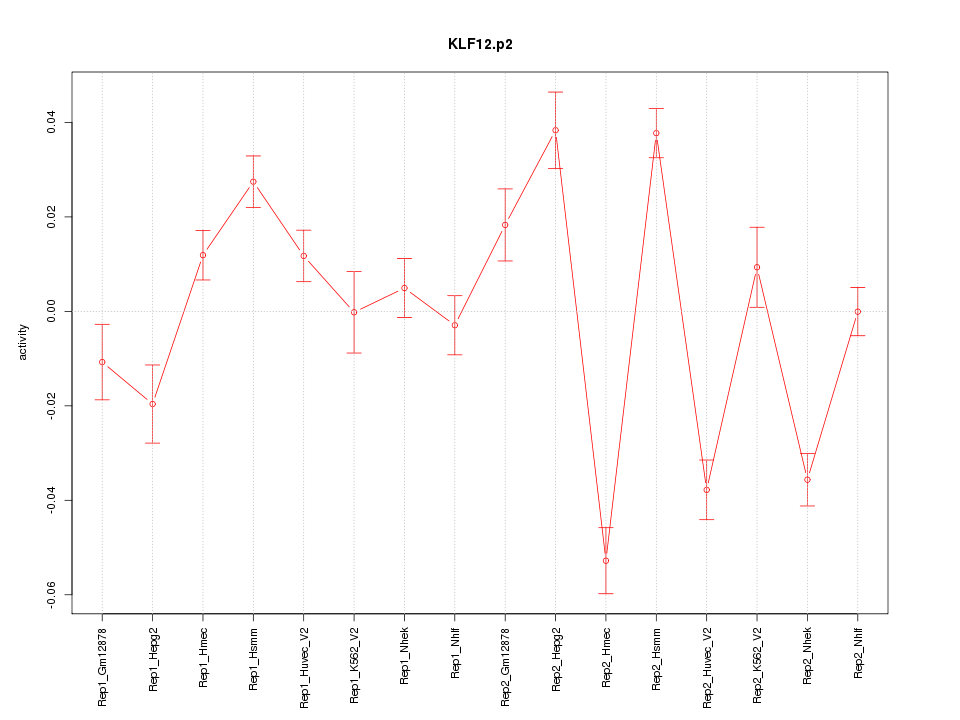
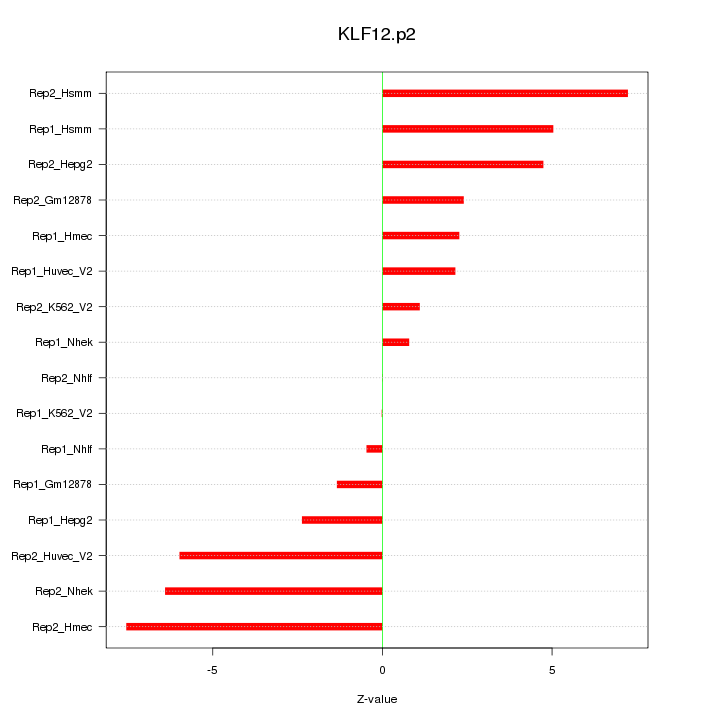
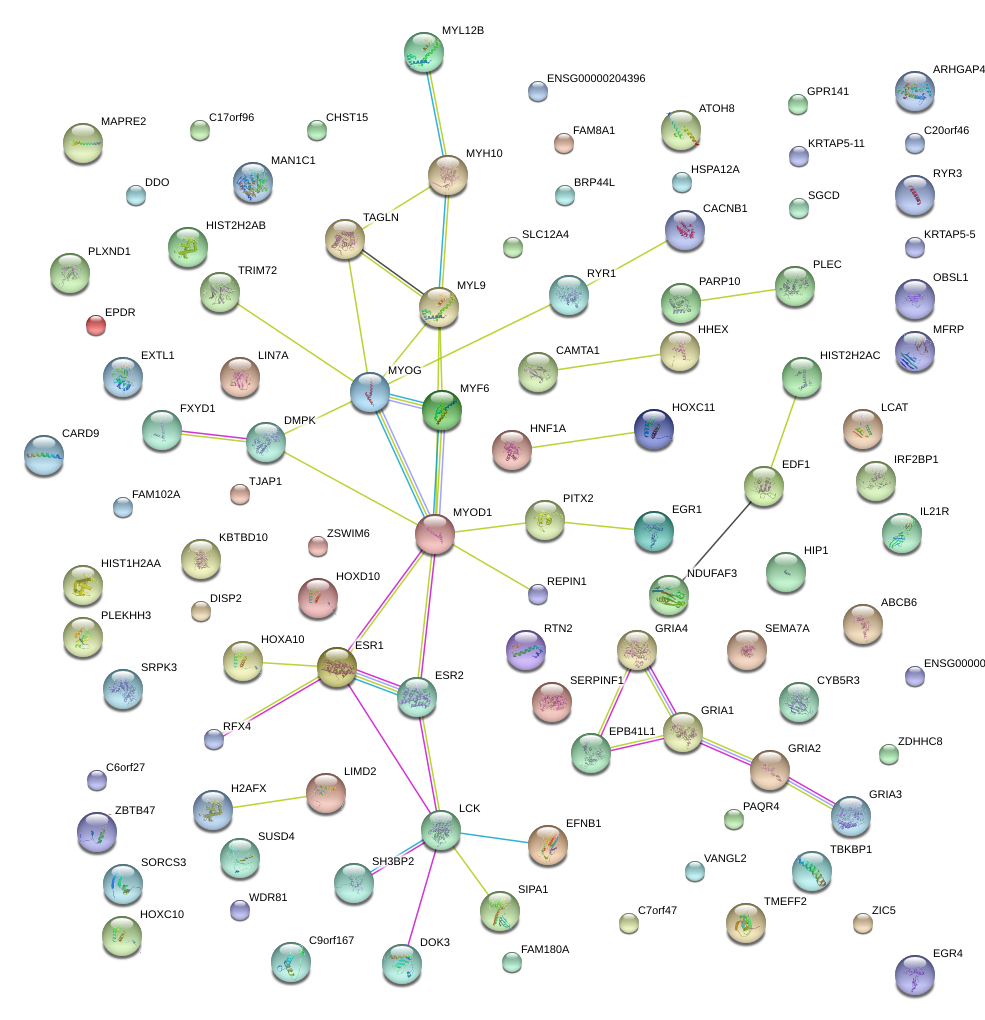
|
chr16_+_31132842
|
3.249
|
NM_001008274
|
TRIM72
|
tripartite motif containing 72
|
|
chr16_-_66535457
|
2.327
|
NM_000229
|
LCAT
|
lecithin-cholesterol acyltransferase
|
|
chr16_-_66535923
|
2.308
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr12_+_52665196
|
2.131
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr12_-_79855580
|
2.097
|
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr2_-_192767494
|
2.079
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr12_-_79855618
|
2.044
|
NM_004664
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr7_-_27180398
|
2.029
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr3_-_130761967
|
2.024
|
|
PLXND1
|
plexin D1
|
|
chr2_+_104837004
|
2.021
|
|
|
|
|
chr17_+_1612092
|
1.983
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr2_+_85834111
|
1.953
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr5_+_152850276
|
1.872
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr5_+_155686333
|
1.872
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr17_+_1612031
|
1.844
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr17_+_1612056
|
1.811
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr10_-_105410840
|
1.781
|
|
|
|
|
chr20_+_34206070
|
1.737
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr2_+_170074457
|
1.722
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr11_+_116575249
|
1.707
|
NM_001001522
NM_003186
|
TAGLN
|
transgelin
|
|
chr17_+_1580504
|
1.682
|
|
WDR81
|
WD repeat domain 81
|
|
chr17_-_8474733
|
1.666
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr1_+_6767968
|
1.648
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr4_+_2783764
|
1.633
|
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr19_-_50975603
|
1.613
|
NM_001081563
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr10_+_106390848
|
1.602
|
NM_014978
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr17_-_34084682
|
1.591
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr16_+_27320983
|
1.579
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr12_+_79625538
|
1.577
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr8_-_145132582
|
1.577
|
NM_032789
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr20_-_1113116
|
1.530
|
NM_018354
|
C20orf46
|
chromosome 20 open reading frame 46
|
|
chr2_-_220144254
|
1.509
|
|
OBSL1
|
obscurin-like 1
|
|
chr5_+_133870175
|
1.487
|
|
|
|
|
chr11_-_118716244
|
1.474
|
|
MFRP
|
membrane frizzled-related protein
|
|
chr1_-_201321711
|
1.421
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr6_+_17708483
|
1.391
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr19_-_50688310
|
1.389
|
NM_206901
|
RTN2
|
reticulon 2
|
|
chr5_+_60663842
|
1.374
|
NM_020928
|
ZSWIM6
|
zinc finger, SWIM-type containing 6
|
|
chr4_-_111777651
|
1.365
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr6_-_166716353
|
1.362
|
|
BRP44L
|
brain protein 44-like
|
|
chr17_-_34607257
|
1.349
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr17_+_71141108
|
1.319
|
NM_001162995
|
LOC643008
|
hypothetical protein LOC643008
|
|
chr4_-_111777584
|
1.318
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr17_+_1611983
|
1.303
|
NM_002615
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr9_-_138880530
|
1.301
|
NM_003792
NM_153200
|
EDF1
|
endothelial differentiation-related factor 1
|
|
chr13_-_99422163
|
1.301
|
NM_033132
|
ZIC5
|
Zic family member 5 (odd-paired homolog, Drosophila)
|
|
chr11_+_17697685
|
1.299
|
NM_002478
|
MYOD1
|
myogenic differentiation 1
|
|
chr10_+_94439658
|
1.296
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr22_-_41372620
|
1.281
|
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chrX_+_152699616
|
1.266
|
NM_001170760
NM_001170761
NM_014370
|
SRPK3
|
SRSF protein kinase 3
|
|
chr7_-_135083906
|
1.256
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr11_-_118471313
|
1.227
|
|
H2AFX
|
H2A histone family, member X
|
|
chr17_+_43127628
|
1.223
|
NM_014726
|
TBKBP1
|
TBK1 binding protein 1
|
|
chr6_+_152168500
|
1.216
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|
|
chr10_-_118491971
|
1.213
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr22_+_18499354
|
1.207
|
NM_001185024
NM_013373
|
ZDHHC8
|
zinc finger, DHHC-type containing 8
|
|
chr7_-_75206152
|
1.203
|
|
HIP1
|
huntingtin interacting protein 1
|
|
chr3_+_49033621
|
1.198
|
NM_199073
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr16_+_2959362
|
1.194
|
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr18_+_30810889
|
1.189
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr20_+_34603311
|
1.173
|
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr11_+_65164767
|
1.173
|
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr7_+_149699185
|
1.173
|
NM_014374
|
REPIN1
|
replication initiator 1
|
|
chr16_+_2959342
|
1.170
|
NM_152341
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr11_-_118471385
|
1.164
|
NM_002105
|
H2AFX
|
H2A histone family, member X
|
|
chr1_+_158636987
|
1.153
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr7_+_37926943
|
1.147
|
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr19_-_50975121
|
1.144
|
|
|
|
|
chr10_+_106391032
|
1.142
|
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr4_-_111763356
|
1.131
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr15_-_72513311
|
1.130
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr9_-_129752687
|
1.130
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr17_-_59131210
|
1.124
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr17_+_12633504
|
1.123
|
NM_014859
|
ARHGAP44
|
Rho GTPase activating protein 44
|
|
chr1_+_26220895
|
1.115
|
|
EXTL1
|
exostoses (multiple)-like 1
|
|
chr10_-_125841840
|
1.112
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr5_+_137829077
|
1.109
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr11_-_558419
|
1.107
|
|
|
|
|
chr3_+_42670179
|
1.103
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr5_-_176869463
|
1.102
|
NM_024872
|
DOK3
|
docking protein 3
|
|
chr17_-_38081967
|
1.101
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr9_-_138387903
|
1.090
|
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chr6_+_43565192
|
1.077
|
NM_001146017
|
TJAP1
|
tight junction associated protein 1 (peripheral)
|
|
chr1_+_25816545
|
1.075
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr19_+_43616167
|
1.070
|
NM_000540
NM_001042723
|
RYR1
|
ryanodine receptor 1 (skeletal)
|
|
chr1_-_221604023
|
1.066
|
NM_001037175
NM_017982
|
SUSD4
|
sushi domain containing 4
|
|
chr19_-_51081147
|
1.063
|
NM_015649
|
IRF2BP1
|
interferon regulatory factor 2 binding protein 1
|
|
chr17_-_59131194
|
1.062
|
|
LIMD2
|
LIM domain containing 2
|
|
chr5_-_176869943
|
1.060
|
NM_001144875
NM_001144876
|
DOK3
|
docking protein 3
|
|
chr5_-_176868483
|
1.057
|
|
DOK3
|
docking protein 3
|
|
chr11_+_1607580
|
1.054
|
NM_001001480
|
KRTAP5-5
|
keratin associated protein 5-5
|
|
chr12_+_119900931
|
1.049
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr19_+_40322231
|
1.045
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr6_-_110843432
|
1.036
|
NM_003649
NM_004032
|
DDO
|
D-aspartate oxidase
|
|
chrX_+_67965517
|
1.030
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr15_+_38437703
|
1.027
|
NM_033510
|
DISP2
|
dispatched homolog 2 (Drosophila)
|
|
chr6_-_31853027
|
1.017
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr2_-_219791902
|
1.012
|
NM_005689
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6
|
|
chr2_-_73373956
|
1.011
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr8_-_145119627
|
1.010
|
NM_201378
|
PLEC
|
plectin
|
|
chr7_-_99871981
|
1.004
|
NM_145030
|
C7orf47
|
chromosome 7 open reading frame 47
|
|
chr1_+_32512298
|
1.003
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr12_+_52653176
|
1.003
|
NM_014212
|
HOXC11
|
homeobox C11
|
|
chr12_+_105519049
|
0.998
|
NM_002920
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr9_+_139292094
|
0.998
|
NM_017723
|
C9orf167
|
chromosome 9 open reading frame 167
|
|
chr19_+_16296734
|
0.997
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr1_-_201321999
|
0.993
|
NM_002479
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr2_-_236741352
|
0.992
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr13_-_78075683
|
0.988
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr7_+_138566770
|
0.982
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr7_+_123083096
|
0.980
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr11_-_118716736
|
0.978
|
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr2_-_73152323
|
0.976
|
NM_144579
|
SFXN5
|
sideroflexin 5
|
|
chr7_+_23496661
|
0.976
|
|
RPS2P32
|
ribosomal protein S2 pseudogene 32
|
|
chr1_+_23990228
|
0.961
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr15_-_72513053
|
0.959
|
NM_001146030
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr1_+_143920424
|
0.957
|
NM_203458
|
NOTCH2NL
|
notch 2 N-terminal like
|
|
chr1_+_201364050
|
0.942
|
|
ADORA1
|
adenosine A1 receptor
|
|
chr2_+_219532590
|
0.938
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr9_-_138387926
|
0.933
|
NM_052813
NM_052814
|
CARD9
|
caspase recruitment domain family, member 9
|
|
chr5_-_149515331
|
0.930
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr17_+_35853177
|
0.926
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr1_-_182273451
|
0.926
|
NM_015101
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr7_-_75461868
|
0.925
|
|
TMEM120A
|
transmembrane protein 120A
|
|
chr5_-_32480586
|
0.925
|
NM_016107
|
ZFR
|
zinc finger RNA binding protein
|
|
chr12_+_119900677
|
0.925
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr21_-_35343448
|
0.924
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr2_+_172658453
|
0.924
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr11_+_65164963
|
0.923
|
|
|
|
|
chr5_+_155686750
|
0.921
|
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr2_+_232281491
|
0.919
|
|
PTMA
|
prothymosin, alpha
|
|
chr5_+_137829065
|
0.918
|
|
EGR1
|
early growth response 1
|
|
chr9_+_138991775
|
0.918
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr6_-_166716449
|
0.917
|
NM_016098
|
BRP44L
|
brain protein 44-like
|
|
chr7_-_75206212
|
0.912
|
NM_005338
|
HIP1
|
huntingtin interacting protein 1
|
|
chr10_+_104394185
|
0.909
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr9_-_103540682
|
0.908
|
NM_133445
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chr1_-_90955365
|
0.907
|
NM_020063
|
BARHL2
|
BarH-like homeobox 2
|
|
chr17_+_23723180
|
0.906
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr19_-_2378530
|
0.903
|
|
TIMM13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr15_+_38550451
|
0.900
|
NM_130468
|
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14
|
|
chr16_+_2503676
|
0.900
|
NM_001198569
NM_001694
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr16_+_2959467
|
0.898
|
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr16_+_2503915
|
0.891
|
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr11_-_1974829
|
0.890
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr21_-_42303517
|
0.888
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr11_-_1974992
|
0.882
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr16_+_7322751
|
0.877
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr17_+_23723220
|
0.877
|
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr7_-_75461814
|
0.876
|
|
TMEM120A
|
transmembrane protein 120A
|
|
chr1_+_63561317
|
0.876
|
NM_012183
|
FOXD3
|
forkhead box D3
|
|
chr4_+_14613395
|
0.874
|
NM_001177381
NM_001177382
NM_001177383
NM_001177384
NM_182485
NM_182646
|
CPEB2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr7_+_101715137
|
0.873
|
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr16_+_2450081
|
0.873
|
NM_025108
|
C16orf59
|
chromosome 16 open reading frame 59
|
|
chr11_-_1599900
|
0.871
|
NM_001012709
|
KRTAP5-4
|
keratin associated protein 5-4
|
|
chr1_-_53380709
|
0.868
|
NM_006671
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7
|
|
chrX_-_152889815
|
0.866
|
NM_005334
|
HCFC1
|
host cell factor C1 (VP16-accessory protein)
|
|
chr12_+_56135353
|
0.865
|
NM_031479
|
INHBE
|
inhibin, beta E
|
|
chr22_-_41353575
|
0.864
|
|
CYB5R3
|
cytochrome b5 reductase 3
|
|
chr1_+_26895108
|
0.864
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr9_-_122728993
|
0.863
|
NM_005658
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr5_-_176869440
|
0.862
|
|
DOK3
|
docking protein 3
|
|
chr1_-_182273114
|
0.861
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr15_+_72005864
|
0.858
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr15_+_72005875
|
0.857
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr2_+_233099113
|
0.854
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr13_+_110637173
|
0.850
|
NM_001113513
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr17_-_1342700
|
0.848
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr8_-_145097031
|
0.843
|
NM_201380
|
PLEC
|
plectin
|
|
chr2_-_74563542
|
0.838
|
|
CCDC142
|
coiled-coil domain containing 142
|
|
chr5_-_16518847
|
0.836
|
|
ZNF622
|
zinc finger protein 622
|
|
chr5_-_16518893
|
0.834
|
NM_033414
|
ZNF622
|
zinc finger protein 622
|
|
chr12_-_69434330
|
0.833
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr21_-_35343456
|
0.830
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr9_+_138991835
|
0.828
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr1_-_38285033
|
0.826
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr16_+_87765661
|
0.822
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr6_-_109882116
|
0.820
|
|
MICAL1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr12_-_69434650
|
0.818
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr14_-_103058568
|
0.817
|
|
CKB
|
creatine kinase, brain
|
|
chr20_+_39199263
|
0.816
|
|
PLCG1
|
phospholipase C, gamma 1
|
|
chr14_-_22891263
|
0.815
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr6_+_11646472
|
0.814
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr1_+_38032045
|
0.813
|
NM_001031740
NM_001113482
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr11_+_66381451
|
0.812
|
NM_024036
|
LRFN4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr9_-_138378061
|
0.809
|
NM_001080849
|
DNLZ
|
DNL-type zinc finger
|
|
chr21_-_35343428
|
0.807
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr15_-_72952648
|
0.807
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr11_-_1725924
|
0.805
|
NM_001170820
|
LOC402778
|
CD225 family protein FLJ76511
|
|
chr15_-_38953628
|
0.800
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr6_+_99389318
|
0.800
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr3_+_50281582
|
0.799
|
|
SEMA3B
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr17_-_7248137
|
0.798
|
NM_152766
|
C17orf61-PLSCR3
C17orf61
|
C17orf61-PLSCR3 read-through transcript
chromosome 17 open reading frame 61
|
|
chr7_+_101715110
|
0.795
|
NM_020979
|
SH2B2
|
SH2B adaptor protein 2
|
|
chr5_-_176857158
|
0.794
|
|
PDLIM7
|
PDZ and LIM domain 7 (enigma)
|
|
chr8_-_145132551
|
0.793
|
|
PARP10
|
poly (ADP-ribose) polymerase family, member 10
|
|
chr20_+_34603290
|
0.793
|
NM_006097
NM_181526
|
MYL9
|
myosin, light chain 9, regulatory
|
|
chr20_-_604443
|
0.792
|
|
|
|
|
chr16_-_2204776
|
0.791
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr3_-_46693902
|
0.791
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|