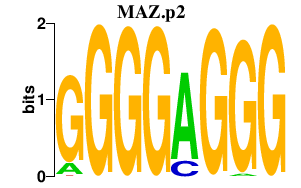
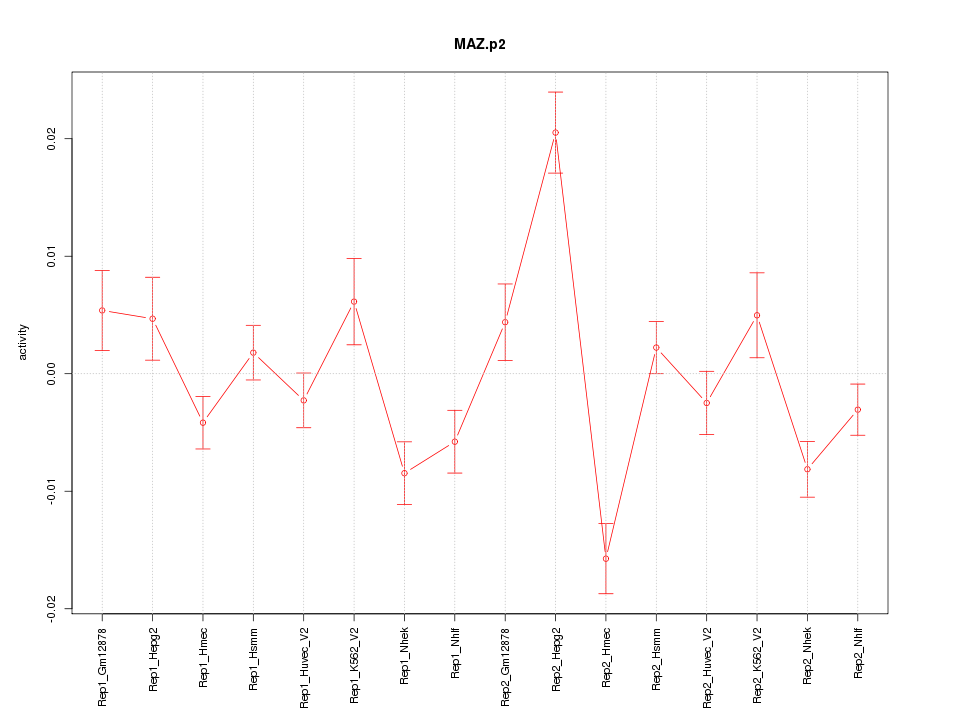
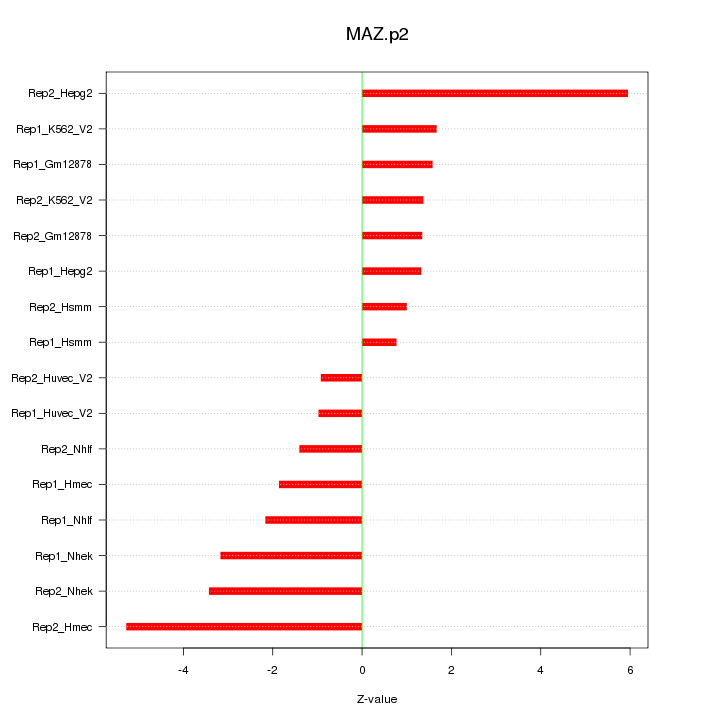
|
chr4_-_809879
|
4.611
|
NM_006651
|
CPLX1
|
complexin 1
|
|
chr8_+_144706699
|
4.486
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chr20_+_61841662
|
4.258
|
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr7_+_5289079
|
4.180
|
NM_001040661
NM_153247
|
SLC29A4
|
solute carrier family 29 (nucleoside transporters), member 4
|
|
chr19_-_55524445
|
4.135
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chr19_+_659766
|
4.087
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chrX_+_138060
|
3.986
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr7_-_158073121
|
3.904
|
NM_002847
NM_130842
NM_130843
|
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr20_+_61841553
|
3.735
|
NM_020062
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr2_-_25328452
|
3.616
|
NM_153759
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr14_-_21064182
|
3.382
|
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr4_-_6253152
|
3.291
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr1_+_26895108
|
3.153
|
NM_006015
NM_139135
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr20_-_34635091
|
2.948
|
|
|
|
|
chr12_+_56291484
|
2.917
|
NM_182947
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chrX_+_9392980
|
2.908
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr1_-_32574185
|
2.905
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr2_+_42128521
|
2.880
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr11_+_56984702
|
2.859
|
NM_178570
|
RTN4RL2
|
reticulon 4 receptor-like 2
|
|
chr19_-_2672337
|
2.779
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr16_+_84202515
|
2.652
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr4_+_995395
|
2.641
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr22_+_48927610
|
2.496
|
NM_001164106
|
MOV10L1
|
Mov10l1, Moloney leukemia virus 10-like 1, homolog (mouse)
|
|
chr16_-_2186465
|
2.483
|
NM_020764
|
CASKIN1
|
CASK interacting protein 1
|
|
chr6_+_99389318
|
2.452
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr5_+_139008209
|
2.425
|
NM_016463
|
CXXC5
|
CXXC finger protein 5
|
|
chr20_-_61755145
|
2.387
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr17_-_39652449
|
2.376
|
NM_001076684
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr13_-_113066366
|
2.299
|
NM_024719
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr9_+_70509925
|
2.267
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr15_-_62854749
|
2.250
|
NM_194272
|
RBPMS2
|
RNA binding protein with multiple splicing 2
|
|
chr18_-_28606839
|
2.250
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr5_-_134397740
|
2.241
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr20_+_34635362
|
2.205
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr9_-_139564656
|
2.181
|
NM_001098537
NM_152286
|
PNPLA7
|
patatin-like phospholipase domain containing 7
|
|
chr13_-_113066202
|
2.180
|
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr2_-_74634569
|
2.168
|
NM_032603
|
LOXL3
|
lysyl oxidase-like 3
|
|
chr14_+_104261578
|
2.149
|
NM_152328
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr20_+_44083667
|
2.117
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr10_-_135021502
|
2.093
|
NM_001098483
NM_198472
|
C10orf125
|
chromosome 10 open reading frame 125
|
|
chr3_-_10522221
|
2.065
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr3_-_10522267
|
2.062
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr19_+_14405216
|
2.054
|
|
PKN1
|
protein kinase N1
|
|
chr17_-_53764867
|
2.031
|
|
|
|
|
chr2_+_72998111
|
2.029
|
NM_004097
|
EMX1
|
empty spiracles homeobox 1
|
|
chr20_-_44370537
|
1.994
|
|
CDH22
|
cadherin 22, type 2
|
|
chr17_-_39652391
|
1.993
|
|
UBTF
|
upstream binding transcription factor, RNA polymerase I
|
|
chr6_+_99389300
|
1.984
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr1_-_149698516
|
1.977
|
NM_001194937
NM_001194938
NM_015100
NM_145796
|
POGZ
|
pogo transposable element with ZNF domain
|
|
chr3_-_10522340
|
1.967
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr22_+_38183480
|
1.960
|
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr3_-_47595190
|
1.956
|
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr16_+_49139694
|
1.949
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr12_+_93066370
|
1.947
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr7_-_102044374
|
1.927
|
|
RASA4
|
RAS p21 protein activator 4
|
|
chr1_+_3679168
|
1.926
|
NM_001163724
|
LOC388588
|
hypothetical protein LOC388588
|
|
chr6_+_7052985
|
1.926
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr17_-_39632845
|
1.923
|
|
ATXN7L3
|
ataxin 7-like 3
|
|
chr2_+_220116986
|
1.910
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr1_-_36562341
|
1.883
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr17_-_36938025
|
1.878
|
|
KRT19
|
keratin 19
|
|
chr11_+_47236072
|
1.856
|
NM_001130101
NM_005693
|
NR1H3
|
nuclear receptor subfamily 1, group H, member 3
|
|
chr2_+_220117009
|
1.851
|
|
TMEM198
|
transmembrane protein 198
|
|
chr17_+_35751796
|
1.827
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr16_-_51138306
|
1.825
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr11_+_46358909
|
1.812
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr6_+_7052798
|
1.804
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr2_-_200030944
|
1.778
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr2_+_149118791
|
1.777
|
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr5_+_139008139
|
1.777
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr19_+_12810258
|
1.771
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr22_+_27931685
|
1.770
|
NM_133455
|
EMID1
|
EMI domain containing 1
|
|
chr13_+_99431417
|
1.765
|
|
|
|
|
chr17_+_72880831
|
1.752
|
NM_001113493
|
SEPT9
|
septin 9
|
|
chr19_+_45728270
|
1.734
|
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr7_-_102044419
|
1.719
|
NM_001079877
NM_006989
|
RASA4
|
RAS p21 protein activator 4
|
|
chr4_+_995609
|
1.715
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr17_-_37586716
|
1.712
|
NM_012285
|
KCNH4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4
|
|
chr9_+_138726737
|
1.708
|
NM_152421
|
FAM69B
|
family with sequence similarity 69, member B
|
|
chr15_-_43267139
|
1.688
|
|
SHF
|
Src homology 2 domain containing F
|
|
chr17_+_45008473
|
1.676
|
|
NXPH3
|
neurexophilin 3
|
|
chr20_+_60918836
|
1.671
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr11_+_279113
|
1.663
|
NM_025092
|
ATHL1
|
ATH1, acid trehalase-like 1 (yeast)
|
|
chr7_-_149101227
|
1.655
|
NM_207336
|
ZNF467
|
zinc finger protein 467
|
|
chr5_+_139008091
|
1.650
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr20_-_3097070
|
1.645
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr14_-_20562962
|
1.631
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr2_+_112372651
|
1.631
|
NM_006343
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr2_+_112372671
|
1.628
|
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chrX_+_118776572
|
1.627
|
NM_001105576
|
ANKRD58
|
ankyrin repeat domain 58
|
|
chr19_-_48835783
|
1.626
|
NM_145296
|
CADM4
|
cell adhesion molecule 4
|
|
chr15_-_73530829
|
1.617
|
NM_015477
|
SIN3A
|
SIN3 homolog A, transcription regulator (yeast)
|
|
chr19_+_45728207
|
1.617
|
NM_025213
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr16_+_87765661
|
1.600
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr7_-_86942910
|
1.576
|
NM_000443
NM_018849
NM_018850
|
ABCB4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4
|
|
chr19_-_2653662
|
1.573
|
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr11_+_359723
|
1.566
|
NM_178537
|
B4GALNT4
|
beta-1,4-N-acetyl-galactosaminyl transferase 4
|
|
chr7_+_150390566
|
1.563
|
|
SLC4A2
|
solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1)
|
|
chr16_+_1979959
|
1.549
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chr17_-_34014390
|
1.545
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr2_+_149119029
|
1.543
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr19_-_772966
|
1.535
|
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr6_+_169866714
|
1.535
|
|
LOC729088
|
hypothetical LOC729088
|
|
chr2_+_232281491
|
1.532
|
|
PTMA
|
prothymosin, alpha
|
|
chr6_+_37245860
|
1.532
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr2_+_74635019
|
1.523
|
NM_001381
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr10_+_124885556
|
1.522
|
NM_001105574
|
HMX3
|
H6 family homeobox 3
|
|
chr2_-_174537354
|
1.518
|
|
SP3
|
Sp3 transcription factor
|
|
chr17_+_27617307
|
1.515
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr19_+_59061422
|
1.501
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr18_+_75261231
|
1.498
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr12_-_47679176
|
1.496
|
NM_015086
|
DDN
|
dendrin
|
|
chr6_+_44346457
|
1.494
|
NM_001137560
|
TMEM151B
|
transmembrane protein 151B
|
|
chr14_+_23508922
|
1.493
|
NM_001193636
NM_001193637
NM_001193635
|
DHRS4L2
DHRS4L1
|
dehydrogenase/reductase (SDR family) member 4 like 2
dehydrogenase/reductase (SDR family) member 4 like 1
|
|
chr10_+_45189617
|
1.485
|
NM_000698
|
ALOX5
|
arachidonate 5-lipoxygenase
|
|
chr16_+_19087035
|
1.484
|
NM_016524
|
SYT17
|
synaptotagmin XVII
|
|
chr1_+_945331
|
1.482
|
NM_198576
|
AGRN
|
agrin
|
|
chr2_+_115635982
|
1.481
|
NM_001178034
NM_001004360
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr12_+_6808025
|
1.476
|
|
LEPREL2
|
leprecan-like 2
|
|
chr19_-_18409788
|
1.475
|
NM_001170939
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr12_+_130945215
|
1.468
|
NM_003565
|
ULK1
|
unc-51-like kinase 1 (C. elegans)
|
|
chr12_+_54700955
|
1.460
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr6_-_32265489
|
1.459
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr16_-_1369613
|
1.454
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr12_-_56418295
|
1.452
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr20_-_49592444
|
1.446
|
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr14_+_23907065
|
1.443
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr12_+_6290359
|
1.428
|
NM_001144856
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr14_+_31615922
|
1.425
|
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr12_+_56230110
|
1.425
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr20_-_22512893
|
1.412
|
|
FOXA2
|
forkhead box A2
|
|
chr19_-_772913
|
1.411
|
NM_024888
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr17_+_45008175
|
1.408
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr21_+_42946719
|
1.405
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr20_-_49592654
|
1.392
|
NM_012340
NM_173091
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr19_-_8581559
|
1.391
|
NM_030957
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10
|
|
chr16_+_51722285
|
1.384
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr10_-_103525646
|
1.384
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr12_-_61614930
|
1.384
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr17_+_50698169
|
1.382
|
|
HLF
|
hepatic leukemia factor
|
|
chr9_+_33807168
|
1.382
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr12_+_56230047
|
1.380
|
|
KIF5A
|
kinesin family member 5A
|
|
chr10_+_124897627
|
1.377
|
NM_005519
|
HMX2
|
H6 family homeobox 2
|
|
chr11_+_47958835
|
1.374
|
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr17_-_34157962
|
1.362
|
NM_007144
|
PCGF2
|
polycomb group ring finger 2
|
|
chrX_+_152643516
|
1.358
|
NM_000033
|
ABCD1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr2_+_232281470
|
1.357
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr2_+_79593567
|
1.355
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr20_+_42807901
|
1.349
|
NM_022358
|
KCNK15
|
potassium channel, subfamily K, member 15
|
|
chr4_+_996251
|
1.348
|
NM_021923
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr19_+_1397268
|
1.345
|
|
APC2
|
adenomatosis polyposis coli 2
|
|
chr12_+_6290127
|
1.339
|
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr7_-_100077072
|
1.338
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr14_+_31615959
|
1.337
|
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr19_-_6453224
|
1.336
|
NM_006087
|
TUBB4
|
tubulin, beta 4
|
|
chr22_+_21742727
|
1.335
|
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr6_+_7053183
|
1.332
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr19_+_40451807
|
1.332
|
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr22_-_40652724
|
1.324
|
NM_052945
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C
|
|
chr17_+_75366524
|
1.321
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr2_+_119905812
|
1.312
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr16_+_2503676
|
1.307
|
NM_001198569
NM_001694
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c
|
|
chr16_+_29730909
|
1.300
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr17_+_72880967
|
1.298
|
|
SEPT9
|
septin 9
|
|
chr5_+_139008667
|
1.297
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr14_-_26136799
|
1.297
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr16_+_1323627
|
1.292
|
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr17_-_71352182
|
1.290
|
NM_199242
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr5_-_134397470
|
1.287
|
|
PITX1
|
paired-like homeodomain 1
|
|
chr1_-_40903897
|
1.286
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr10_+_94439658
|
1.285
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr17_-_33982580
|
1.277
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr7_-_51351952
|
1.275
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr14_-_21074875
|
1.271
|
NM_005407
|
SALL2
|
sal-like 2 (Drosophila)
|
|
chr14_+_20608179
|
1.269
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chr1_+_39319675
|
1.268
|
NM_012090
|
MACF1
|
microtubule-actin crosslinking factor 1
|
|
chr6_+_163755788
|
1.264
|
|
QKI
|
quaking homolog, KH domain RNA binding (mouse)
|
|
chr12_-_47679459
|
1.254
|
|
DDN
|
dendrin
|
|
chr2_-_25418249
|
1.251
|
NM_022552
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha
|
|
chr17_+_16259812
|
1.247
|
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr10_-_104168890
|
1.246
|
NM_002779
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr15_+_38550451
|
1.245
|
NM_130468
|
CHST14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14
|
|
chr19_+_3310138
|
1.243
|
NM_205843
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr17_-_59131194
|
1.241
|
|
LIMD2
|
LIM domain containing 2
|
|
chr11_-_64268841
|
1.236
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr17_-_59131210
|
1.232
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr19_-_14062230
|
1.225
|
NM_138352
|
SAMD1
|
sterile alpha motif domain containing 1
|
|
chr19_+_8180179
|
1.224
|
NM_024552
|
LASS4
|
LAG1 homolog, ceramide synthase 4
|
|
chr11_+_817584
|
1.219
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr16_+_65196125
|
1.215
|
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3
|
|
chr19_-_2653738
|
1.215
|
NM_052847
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr14_-_37134191
|
1.215
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr2_-_200030412
|
1.209
|
|
SATB2
|
SATB homeobox 2
|
|
chr16_-_30042030
|
1.207
|
|
MAPK3
|
mitogen-activated protein kinase 3
|
|
chr19_-_44060729
|
1.206
|
NM_001195833
NM_198445
|
RINL
|
Ras and Rab interactor-like
|
|
chr6_-_90178375
|
1.202
|
|
RRAGD
|
Ras-related GTP binding D
|
|
chr8_+_11599119
|
1.198
|
NM_002052
|
GATA4
|
GATA binding protein 4
|
|
chr10_+_22650145
|
1.198
|
|
BMI1
|
BMI1 polycomb ring finger oncogene
|
|
chr17_+_44429748
|
1.185
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr4_-_1156533
|
1.183
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|