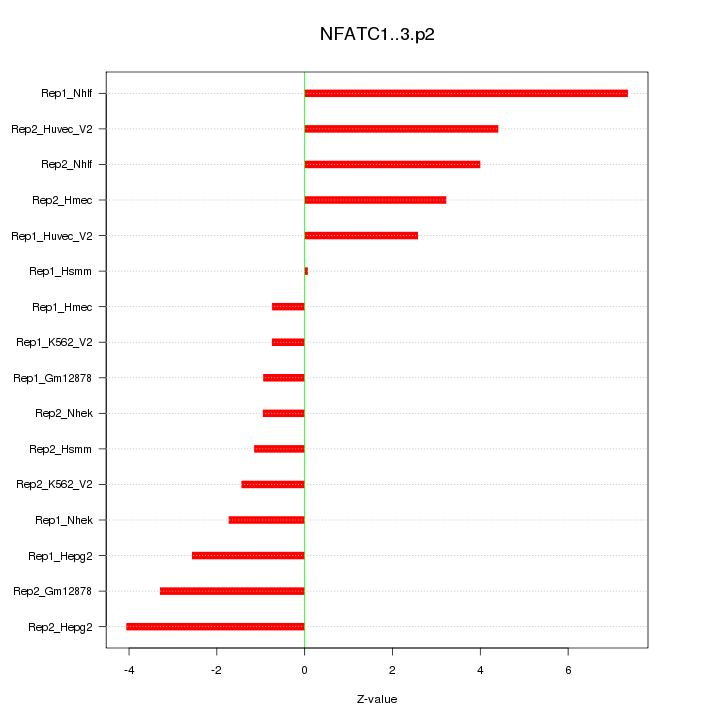
|
chr12_+_13240983
|
9.820
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_+_13240987
|
9.616
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_+_13240868
|
8.266
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr18_-_51219880
|
6.854
|
|
TCF4
|
transcription factor 4
|
|
chr22_-_34566210
|
6.143
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr8_-_13416554
|
5.603
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_160868932
|
5.258
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr1_+_161305558
|
5.100
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr22_-_34566367
|
5.045
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr22_-_34566276
|
5.008
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr18_+_64616296
|
4.809
|
NM_024781
|
CCDC102B
|
coiled-coil domain containing 102B
|
|
chr1_+_160868822
|
4.642
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr1_+_157246305
|
4.507
|
NM_005531
|
IFI16
|
interferon, gamma-inducible protein 16
|
|
chr21_-_27261276
|
4.378
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr12_-_90097384
|
4.279
|
NM_133503
|
DCN
|
decorin
|
|
chr8_-_93144039
|
4.065
|
|
|
|
|
chr2_-_237987545
|
3.955
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_237987553
|
3.790
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_+_66519949
|
3.654
|
|
MEIS1
|
Meis homeobox 1
|
|
chr20_+_42644633
|
3.649
|
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma
|
|
chr8_-_93144366
|
3.515
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_-_36266930
|
3.372
|
NM_001128227
NM_001190388
|
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase
|
|
chr4_-_152366970
|
3.341
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr11_-_85115105
|
3.333
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr13_-_32822759
|
3.292
|
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_+_189547377
|
3.276
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr6_+_39868770
|
3.260
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr9_+_35528890
|
3.209
|
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr2_-_237987509
|
3.181
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr8_-_6407882
|
3.094
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr2_-_237987554
|
3.048
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_-_68470585
|
3.032
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr1_+_66772393
|
2.998
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr2_-_237987579
|
2.943
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr4_-_115120251
|
2.930
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr9_+_74956494
|
2.916
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr5_-_54317102
|
2.881
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr14_+_62740878
|
2.880
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr4_-_153493133
|
2.872
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr2_+_189547323
|
2.794
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_+_118100977
|
2.788
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr17_+_44643067
|
2.788
|
|
ABI3
|
ABI family, member 3
|
|
chr1_-_79244948
|
2.775
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr12_+_32578513
|
2.751
|
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr1_-_149047329
|
2.746
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr3_+_158637273
|
2.704
|
NM_002852
|
PTX3
|
pentraxin 3, long
|
|
chr2_+_189547358
|
2.689
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chrX_+_56772391
|
2.597
|
|
LOC550643
|
hypothetical LOC550643
|
|
chr3_-_113841448
|
2.573
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_+_120416680
|
2.559
|
NM_001105533
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr3_-_150533948
|
2.553
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr19_+_46417117
|
2.539
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr2_-_188021117
|
2.538
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr16_-_63713382
|
2.522
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr16_+_1524162
|
2.505
|
NM_024600
|
TMEM204
|
transmembrane protein 204
|
|
chr9_+_35528628
|
2.432
|
NM_001135999
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr2_-_134042500
|
2.380
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr4_-_55686503
|
2.367
|
NM_002253
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chr16_-_63713485
|
2.363
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr10_-_31328426
|
2.343
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr6_+_116939500
|
2.325
|
NM_153711
|
FAM26E
|
family with sequence similarity 26, member E
|
|
chr16_-_63713466
|
2.308
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr2_-_189752575
|
2.303
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr6_-_134680888
|
2.302
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr21_-_26867138
|
2.293
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr13_-_22905822
|
2.289
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr15_-_40537002
|
2.260
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr4_-_159312973
|
2.252
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr17_+_29606408
|
2.245
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr13_-_79810972
|
2.244
|
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr12_-_14929991
|
2.230
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr1_+_169421011
|
2.205
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr15_+_37330176
|
2.201
|
NM_207445
|
C15orf54
|
chromosome 15 open reading frame 54
|
|
chrX_+_100692071
|
2.199
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr6_-_112682401
|
2.181
|
|
LAMA4
|
laminin, alpha 4
|
|
chr7_-_41709191
|
2.167
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr4_-_141293572
|
2.159
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr8_+_70567634
|
2.103
|
|
SULF1
|
sulfatase 1
|
|
chr6_-_169395847
|
2.101
|
|
THBS2
|
thrombospondin 2
|
|
chr5_+_140742585
|
2.088
|
NM_018920
NM_032087
|
PCDHGA7
|
protocadherin gamma subfamily A, 7
|
|
chr8_+_70567581
|
2.086
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr12_-_48902692
|
2.072
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr11_+_65027721
|
2.053
|
|
|
|
|
chr11_+_113672407
|
2.053
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr6_-_112682459
|
2.032
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr1_+_98899734
|
2.023
|
NM_015976
NM_152238
|
SNX7
|
sorting nexin 7
|
|
chr11_-_68537243
|
2.020
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr5_-_58607673
|
1.975
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_201862845
|
1.940
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr9_+_22437353
|
1.937
|
|
DMRTA1
|
DMRT-like family A1
|
|
chr6_-_112186977
|
1.927
|
NM_153048
|
FYN
|
FYN oncogene related to SRC, FGR, YES
|
|
chr4_+_71806538
|
1.924
|
NM_001037442
NM_014961
|
RUFY3
|
RUN and FYVE domain containing 3
|
|
chr6_-_112682573
|
1.885
|
|
LAMA4
|
laminin, alpha 4
|
|
chr9_-_35679902
|
1.878
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr6_-_76259976
|
1.854
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr7_+_93388951
|
1.840
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr1_+_78243181
|
1.837
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr3_+_25444753
|
1.834
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr13_+_96672541
|
1.828
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr2_-_198248828
|
1.828
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr3_+_25445071
|
1.828
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr11_-_35503720
|
1.808
|
NM_001001991
NM_015430
|
PAMR1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr9_-_90983195
|
1.808
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr6_-_169395972
|
1.804
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr7_+_22733626
|
1.789
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr2_-_202271598
|
1.782
|
NM_033066
|
MPP4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4)
|
|
chr9_-_35679799
|
1.780
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr3_+_45042754
|
1.771
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr22_+_29808845
|
1.762
|
|
SMTN
|
smoothelin
|
|
chr4_-_152368492
|
1.756
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr5_-_59100091
|
1.751
|
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_44642934
|
1.750
|
NM_001198754
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr17_-_44022595
|
1.736
|
|
HOXB3
|
homeobox B3
|
|
chr19_-_48401532
|
1.732
|
NM_002780
NM_213633
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4
|
|
chr10_-_116434364
|
1.725
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr4_-_149582948
|
1.720
|
NM_000901
NM_001166104
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr12_-_21985433
|
1.709
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr1_+_78888064
|
1.703
|
NM_006417
|
IFI44
|
interferon-induced protein 44
|
|
chr12_+_12829807
|
1.703
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr2_-_231697999
|
1.698
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr2_+_33514896
|
1.689
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr3_-_113842553
|
1.672
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_+_158743852
|
1.661
|
NM_001099777
NM_001130001
NM_001130002
|
C3orf55
|
chromosome 3 open reading frame 55
|
|
chr6_+_10663934
|
1.648
|
NM_001491
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group)
|
|
chr12_+_2032964
|
1.638
|
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr4_+_134289893
|
1.637
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr3_-_113842652
|
1.627
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr11_+_48303047
|
1.622
|
NM_001004702
|
OR4C3
|
olfactory receptor, family 4, subfamily C, member 3
|
|
chr9_-_128924819
|
1.614
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chr1_+_183970093
|
1.612
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr2_-_227952249
|
1.612
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr6_-_152744022
|
1.611
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr5_-_59100194
|
1.606
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_-_192419896
|
1.603
|
|
SDPR
|
serum deprivation response
|
|
chr17_-_64649608
|
1.574
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr11_-_71458739
|
1.573
|
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr5_+_38481392
|
1.567
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr5_+_140187746
|
1.551
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr4_+_95595419
|
1.545
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr14_+_85066265
|
1.540
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr9_+_12764988
|
1.540
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr6_-_56824371
|
1.540
|
|
DST
|
dystonin
|
|
chr18_+_7936885
|
1.540
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr2_-_224974954
|
1.540
|
NM_001122779
NM_024785
|
FAM124B
|
family with sequence similarity 124B
|
|
chr3_-_143230124
|
1.534
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr19_+_42689881
|
1.519
|
|
ZNF793
|
zinc finger protein 793
|
|
chr6_+_106640903
|
1.519
|
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr11_+_65023556
|
1.519
|
|
|
|
|
chr17_-_30783631
|
1.508
|
NM_018042
|
SLFN12
|
schlafen family member 12
|
|
chr7_+_136204371
|
1.499
|
NM_000739
NM_001006631
NM_001006632
NM_001006626
NM_001006629
NM_001006628
|
CHRM2
|
cholinergic receptor, muscarinic 2
|
|
chr2_-_133144950
|
1.490
|
NM_144586
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr5_-_146813343
|
1.485
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr1_+_168898898
|
1.475
|
|
PRRX1
|
paired related homeobox 1
|
|
chr4_+_169654742
|
1.475
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr13_-_37070862
|
1.458
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr5_+_155686750
|
1.458
|
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr15_-_50316996
|
1.456
|
|
MYO5C
|
myosin VC
|
|
chr15_+_22652790
|
1.451
|
NM_022805
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr3_-_113841200
|
1.443
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_-_94475504
|
1.429
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr7_+_91762227
|
1.428
|
|
|
|
|
chr5_-_124108672
|
1.412
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr5_+_140733664
|
1.405
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr6_+_106653429
|
1.400
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr19_+_46417099
|
1.394
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr8_-_116750427
|
1.394
|
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr5_-_150306298
|
1.393
|
|
ZNF300P1
|
zinc finger protein 300 pseudogene 1
|
|
chr8_-_6680429
|
1.391
|
NM_207411
|
XKR5
|
XK, Kell blood group complex subunit-related family, member 5
|
|
chr2_-_192420168
|
1.391
|
NM_004657
|
SDPR
|
serum deprivation response
|
|
chr21_+_25856311
|
1.387
|
|
MIR155HG
|
MIR155 host gene (non-protein coding)
|
|
chrX_-_43717864
|
1.384
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr1_+_19807168
|
1.373
|
|
|
|
|
chr4_-_90978469
|
1.369
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr7_-_15692551
|
1.367
|
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr4_+_156899575
|
1.366
|
NM_000857
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr5_+_140154627
|
1.364
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr6_-_128883125
|
1.355
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr8_+_107807415
|
1.354
|
NM_001198534
NM_001198535
|
OXR1
|
oxidation resistance 1
|
|
chr9_-_116920232
|
1.347
|
NM_002160
|
TNC
|
tenascin C
|
|
chr22_-_43986896
|
1.347
|
|
KIAA0930
|
KIAA0930
|
|
chr20_-_10148137
|
1.342
|
|
LOC100131208
|
similar to hCG2045825
|
|
chr16_-_54466709
|
1.341
|
NM_001143685
NM_145024
|
CES5A
|
carboxylesterase 5A
|
|
chr8_+_144802972
|
1.338
|
NM_014789
|
ZNF623
|
zinc finger protein 623
|
|
chr13_+_100902942
|
1.334
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr21_-_38792173
|
1.330
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr9_+_74956601
|
1.327
|
|
ANXA1
|
annexin A1
|
|
chr4_-_87593306
|
1.325
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr3_+_69895651
|
1.320
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr13_-_101852028
|
1.318
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr5_+_140714951
|
1.315
|
NM_018917
NM_032053
|
PCDHGA4
|
protocadherin gamma subfamily A, 4
|
|
chr4_-_90197140
|
1.312
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr1_-_89261021
|
1.310
|
NM_018284
|
GBP3
|
guanylate binding protein 3
|
|
chr12_+_77782925
|
1.309
|
|
SYT1
|
synaptotagmin I
|
|
chr12_-_50871953
|
1.309
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr17_+_44642587
|
1.299
|
NM_001135186
NM_016428
|
ABI3
|
ABI family, member 3
|
|
chr10_-_116434102
|
1.297
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr6_-_56824672
|
1.287
|
|
DST
|
dystonin
|
|
chr7_-_13992351
|
1.284
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr1_-_109201228
|
1.284
|
NM_152763
|
AKNAD1
|
AKNA domain containing 1
|
|
chr11_+_101488380
|
1.282
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|