|
chr7_+_17304707
|
8.475
|
NM_001621
|
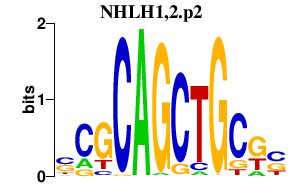
AHR
|
aryl hydrocarbon receptor
|
|
chr2_+_109112428
|
7.674
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr3_-_73756668
|
6.745
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr9_-_83493415
|
5.984
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr9_+_17124974
|
5.741
|
NM_001114395
NM_017738
|
CNTLN
|
centlein, centrosomal protein
|
|
chr2_-_229844198
|
5.410
|
NM_001100818
NM_017933
|
PID1
|
phosphotyrosine interaction domain containing 1
|
|
chr13_-_25523164
|
4.977
|
NM_001007538
|
SHISA2
|
shisa homolog 2 (Xenopus laevis)
|
|
chr5_+_129268352
|
4.308
|
NM_175856
|
CHSY3
|
chondroitin sulfate synthase 3
|
|
chr7_+_55054416
|
3.806
|
|
EGFR
|
epidermal growth factor receptor
|
|
chr6_+_133604203
|
3.801
|
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr19_+_10597167
|
3.774
|
NM_020428
|
SLC44A2
|
solute carrier family 44, member 2
|
|
chr22_-_22652018
|
3.625
|
NM_001355
|
DDT
|
D-dopachrome tautomerase
|
|
chr12_-_74710741
|
3.623
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr7_-_45927263
|
3.604
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr11_-_6298284
|
3.527
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr8_-_24828134
|
3.411
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr1_+_77520315
|
3.381
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr13_-_43259032
|
3.361
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr12_+_51730101
|
3.269
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr3_-_87122936
|
3.261
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr7_-_84654106
|
3.258
|
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr9_-_21549696
|
3.256
|
|
LOC554202
|
hypothetical LOC554202
|
|
chr15_+_90738108
|
3.247
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr12_-_29827952
|
3.206
|
NM_001193451
|
TMTC1
|
transmembrane and tetratricopeptide repeat containing 1
|
|
chr3_-_87122958
|
3.178
|
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr6_+_129245978
|
3.105
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr3_-_31997738
|
3.082
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr12_+_64504835
|
3.069
|
|
HMGA2
|
high mobility group AT-hook 2
|
|
chr13_-_35603466
|
3.062
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr5_-_58370693
|
3.019
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_+_158216631
|
2.994
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr3_-_30911156
|
2.979
|
NM_207359
|
GADL1
|
glutamate decarboxylase-like 1
|
|
chr4_-_40911212
|
2.945
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr14_+_22375753
|
2.876
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr13_-_35603432
|
2.863
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr9_-_35679799
|
2.833
|
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr13_-_35603513
|
2.806
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr1_-_199704773
|
2.790
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr7_+_129919330
|
2.782
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr5_-_136862130
|
2.745
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr13_-_71338144
|
2.718
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr13_-_95094920
|
2.670
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr8_-_16903911
|
2.664
|
NM_019851
|
FGF20
|
fibroblast growth factor 20
|
|
chr10_-_15801661
|
2.660
|
NM_003638
|
ITGA8
|
integrin, alpha 8
|
|
chr19_-_50977485
|
2.658
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr9_-_35679902
|
2.645
|
NM_003289
NM_213674
|
TPM2
|
tropomyosin 2 (beta)
|
|
chr2_-_192767844
|
2.605
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr9_-_25668230
|
2.575
|
|
TUSC1
|
tumor suppressor candidate 1
|
|
chr11_+_110916629
|
2.569
|
|
LAYN
|
layilin
|
|
chr18_-_53621277
|
2.560
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr18_+_11679135
|
2.556
|
NM_182978
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr1_-_59020875
|
2.507
|
|
JUN
|
jun proto-oncogene
|
|
chr7_-_138371310
|
2.486
|
NM_080660
|
ZC3HAV1L
|
zinc finger CCCH-type, antiviral 1-like
|
|
chr2_-_19421852
|
2.475
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr1_+_238321794
|
2.410
|
NM_020066
|
FMN2
|
formin 2
|
|
chr10_-_125841840
|
2.385
|
NM_014863
NM_015892
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr8_+_97575370
|
2.385
|
|
SDC2
|
syndecan 2
|
|
chr20_+_13924263
|
2.346
|
|
MACROD2
|
MACRO domain containing 2
|
|
chr5_-_54317102
|
2.339
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr14_+_37748345
|
2.319
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr3_+_11171213
|
2.314
|
NM_001098212
|
HRH1
|
histamine receptor H1
|
|
chr6_-_82518576
|
2.309
|
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chrX_+_12066505
|
2.309
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr2_+_29191711
|
2.303
|
NM_024692
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4
|
|
chr7_-_32076985
|
2.294
|
NM_001191056
NM_001191057
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa
|
|
chr11_+_7997366
|
2.287
|
|
TUB
|
tubby homolog (mouse)
|
|
chr10_+_112247614
|
2.276
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr17_+_56832492
|
2.276
|
|
TBX2
|
T-box 2
|
|
chr2_-_227371698
|
2.274
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chrX_-_106846253
|
2.251
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr11_+_66827494
|
2.229
|
NM_017857
|
SSH3
|
slingshot homolog 3 (Drosophila)
|
|
chr2_+_149895274
|
2.228
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr8_-_125809559
|
2.216
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr3_-_147361465
|
2.193
|
|
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr10_+_124211353
|
2.183
|
|
HTRA1
|
HtrA serine peptidase 1
|
|
chr6_+_17389752
|
2.167
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr12_-_47468985
|
2.166
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chrX_-_153767625
|
2.157
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr14_-_51605487
|
2.151
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr8_+_1937711
|
2.147
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr9_-_71476917
|
2.129
|
NM_001163
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1
|
|
chr1_-_33420213
|
2.128
|
NM_018207
|
TRIM62
|
tripartite motif containing 62
|
|
chr19_-_1083109
|
2.113
|
NM_001100122
|
SBNO2
|
strawberry notch homolog 2 (Drosophila)
|
|
chr14_-_51605461
|
2.072
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr6_-_153494076
|
2.070
|
NM_012419
|
RGS17
|
regulator of G-protein signaling 17
|
|
chr3_-_79151232
|
2.063
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr3_+_30623376
|
2.043
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr11_+_66827563
|
2.035
|
|
SSH3
|
slingshot homolog 3 (Drosophila)
|
|
chr4_+_7245218
|
2.021
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr11_-_40272239
|
2.017
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr14_-_60817709
|
2.000
|
|
TMEM30B
|
transmembrane protein 30B
|
|
chr2_+_5750229
|
1.994
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr14_-_60817621
|
1.988
|
|
|
|
|
chr11_+_86189138
|
1.986
|
NM_007173
|
PRSS23
|
protease, serine, 23
|
|
chr9_-_21999270
|
1.984
|
NM_004936
NM_078487
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4)
|
|
chr1_+_199519202
|
1.950
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr2_+_238060602
|
1.946
|
NM_001042467
NM_024101
|
MLPH
|
melanophilin
|
|
chr9_+_71848555
|
1.945
|
|
MAMDC2
|
MAM domain containing 2
|
|
chr9_-_90983195
|
1.944
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr1_-_85703321
|
1.943
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr22_+_22652248
|
1.928
|
NM_000854
|
GSTT2
GSTT2B
|
glutathione S-transferase theta 2
glutathione S-transferase theta 2B (gene/pseudogene)
|
|
chr17_+_67628772
|
1.924
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr6_+_142664490
|
1.919
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr11_+_12355600
|
1.909
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr15_+_45797977
|
1.903
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr1_+_77520190
|
1.896
|
|
AK5
|
adenylate kinase 5
|
|
chr6_-_105691675
|
1.869
|
|
BVES
|
blood vessel epicardial substance
|
|
chr1_+_208472817
|
1.857
|
NM_019605
|
SERTAD4
|
SERTA domain containing 4
|
|
chr17_-_15495534
|
1.849
|
|
TRIM16
|
tripartite motif containing 16
|
|
chr8_+_97575244
|
1.846
|
|
SDC2
|
syndecan 2
|
|
chr14_-_51605515
|
1.840
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr4_-_86106567
|
1.833
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr1_-_95165089
|
1.831
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr17_-_19711580
|
1.823
|
NM_001142610
NM_014683
|
ULK2
|
unc-51-like kinase 2 (C. elegans)
|
|
chr5_-_178704934
|
1.810
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chrX_-_99551926
|
1.809
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr1_+_15145001
|
1.795
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr6_+_3696224
|
1.778
|
|
|
|
|
chr5_+_72957736
|
1.775
|
NM_001080479
NM_001177693
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr17_+_56832255
|
1.774
|
|
TBX2
|
T-box 2
|
|
chr1_+_31658549
|
1.772
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr20_+_13150417
|
1.770
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr8_-_11096257
|
1.766
|
NM_173683
|
XKR6
|
XK, Kell blood group complex subunit-related family, member 6
|
|
chr1_+_68070573
|
1.764
|
|
|
|
|
chr13_-_76358370
|
1.759
|
|
KCTD12
|
potassium channel tetramerisation domain containing 12
|
|
chr2_+_191818124
|
1.752
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr13_-_73605914
|
1.743
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr15_+_72006015
|
1.742
|
|
LOXL1
|
lysyl oxidase-like 1
|
|
chr4_+_61749540
|
1.732
|
|
LPHN3
|
latrophilin 3
|
|
chr15_-_53996597
|
1.724
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr9_+_111582317
|
1.721
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr21_+_45318861
|
1.720
|
NM_001112
NM_001160230
NM_015833
NM_015834
|
ADARB1
|
adenosine deaminase, RNA-specific, B1
|
|
chr4_+_37569093
|
1.719
|
NM_015173
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr13_-_71338558
|
1.719
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chrX_-_13866451
|
1.714
|
|
GPM6B
|
glycoprotein M6B
|
|
chr3_+_185538998
|
1.709
|
|
FAM131A
|
family with sequence similarity 131, member A
|
|
chr6_-_128883125
|
1.700
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chrX_+_51092407
|
1.697
|
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10
|
|
chrX_+_73557809
|
1.691
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr14_+_22375747
|
1.678
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr19_-_50977586
|
1.675
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr8_+_97575196
|
1.666
|
|
SDC2
|
syndecan 2
|
|
chr14_+_37748031
|
1.662
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr2_-_230287442
|
1.660
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr7_-_27136876
|
1.657
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr5_-_136862317
|
1.648
|
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr22_+_44277354
|
1.647
|
NM_001996
NM_006485
NM_006486
NM_006487
|
FBLN1
|
fibulin 1
|
|
chr4_-_55686503
|
1.615
|
NM_002253
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chr11_+_113435640
|
1.613
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr7_+_129919134
|
1.607
|
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr4_+_159311353
|
1.588
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr14_+_99181232
|
1.586
|
NM_001127258
NM_032425
|
HHIPL1
|
HHIP-like 1
|
|
chr3_+_100840129
|
1.582
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr17_-_36436979
|
1.573
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr19_-_50977592
|
1.572
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr7_+_129919157
|
1.572
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr16_+_48657381
|
1.558
|
NM_182922
|
HEATR3
|
HEAT repeat containing 3
|
|
chr7_+_27102508
|
1.555
|
|
|
|
|
chr8_-_82186832
|
1.554
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr3_+_49034050
|
1.551
|
NM_199069
NM_199070
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
|
|
chr3_-_62836052
|
1.550
|
NM_003716
NM_183393
NM_183394
|
CADPS
|
Ca++-dependent secretion activator
|
|
chr8_+_24827181
|
1.536
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr12_+_50587468
|
1.533
|
NM_000020
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr13_-_22905822
|
1.532
|
NM_014363
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin)
|
|
chr8_-_41286088
|
1.530
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr11_+_106967283
|
1.523
|
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr5_+_74197473
|
1.523
|
|
LOC441086
|
hypothetical LOC441086
|
|
chr1_-_56817717
|
1.521
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr14_-_37795290
|
1.516
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr10_+_72642274
|
1.513
|
NM_170744
|
UNC5B
|
unc-5 homolog B (C. elegans)
|
|
chr20_+_19903778
|
1.512
|
|
RIN2
|
Ras and Rab interactor 2
|
|
chr6_-_105691235
|
1.504
|
NM_007073
|
BVES
|
blood vessel epicardial substance
|
|
chr5_+_76150588
|
1.504
|
NM_005242
|
F2RL1
|
coagulation factor II (thrombin) receptor-like 1
|
|
chr9_+_133154896
|
1.503
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr4_-_57671307
|
1.499
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr7_+_29200850
|
1.492
|
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr5_+_159276317
|
1.488
|
NM_000679
|
ADRA1B
|
adrenergic, alpha-1B-, receptor
|
|
chr2_-_216944911
|
1.485
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr6_-_46246543
|
1.479
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr12_+_6179736
|
1.477
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr9_-_85342868
|
1.467
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr7_+_98084531
|
1.466
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr10_+_99463446
|
1.465
|
NM_031484
|
MARVELD1
|
MARVEL domain containing 1
|
|
chr20_+_13924014
|
1.465
|
NM_080676
|
MACROD2
|
MACRO domain containing 2
|
|
chr6_-_72187168
|
1.463
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr13_-_73606362
|
1.459
|
|
KLF12
|
Kruppel-like factor 12
|
|
chr9_-_16860664
|
1.456
|
|
BNC2
|
basonuclin 2
|
|
chr17_-_7434113
|
1.448
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr6_+_133604181
|
1.445
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr13_+_112746908
|
1.444
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr13_-_51878235
|
1.439
|
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr9_-_139060416
|
1.439
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr1_+_13782838
|
1.438
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr2_-_133144950
|
1.438
|
NM_144586
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr14_-_60817663
|
1.435
|
|
TMEM30B
|
transmembrane protein 30B
|
|
chr8_+_24827173
|
1.424
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr14_+_22375656
|
1.420
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr5_+_92946348
|
1.419
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr11_+_12987455
|
1.419
|
NM_001080521
|
RASSF10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10
|
|
chr14_+_62740878
|
1.418
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|