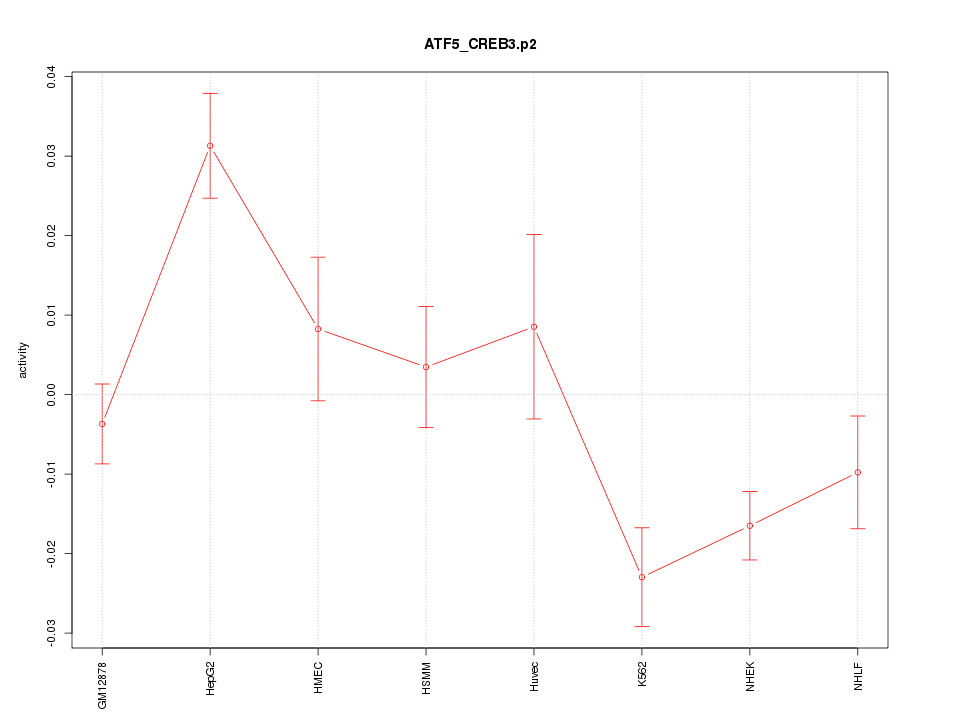
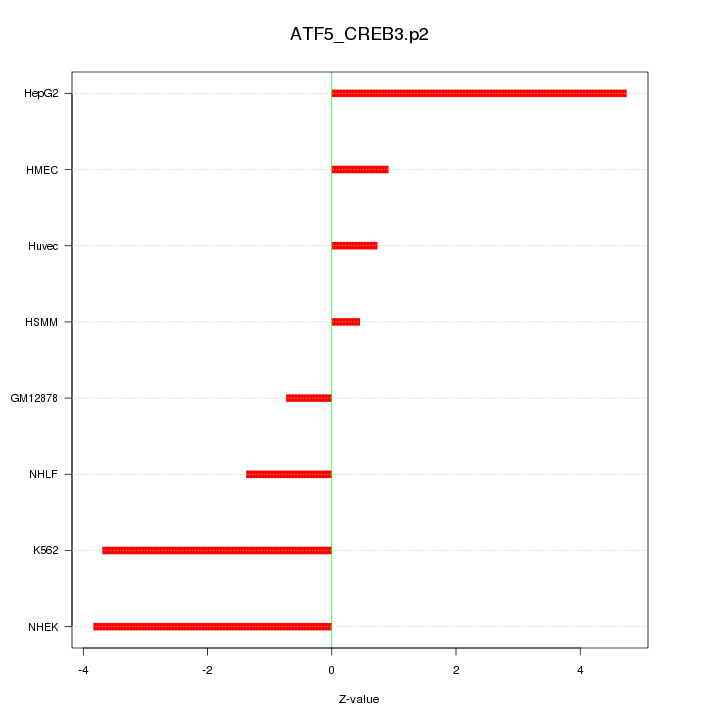
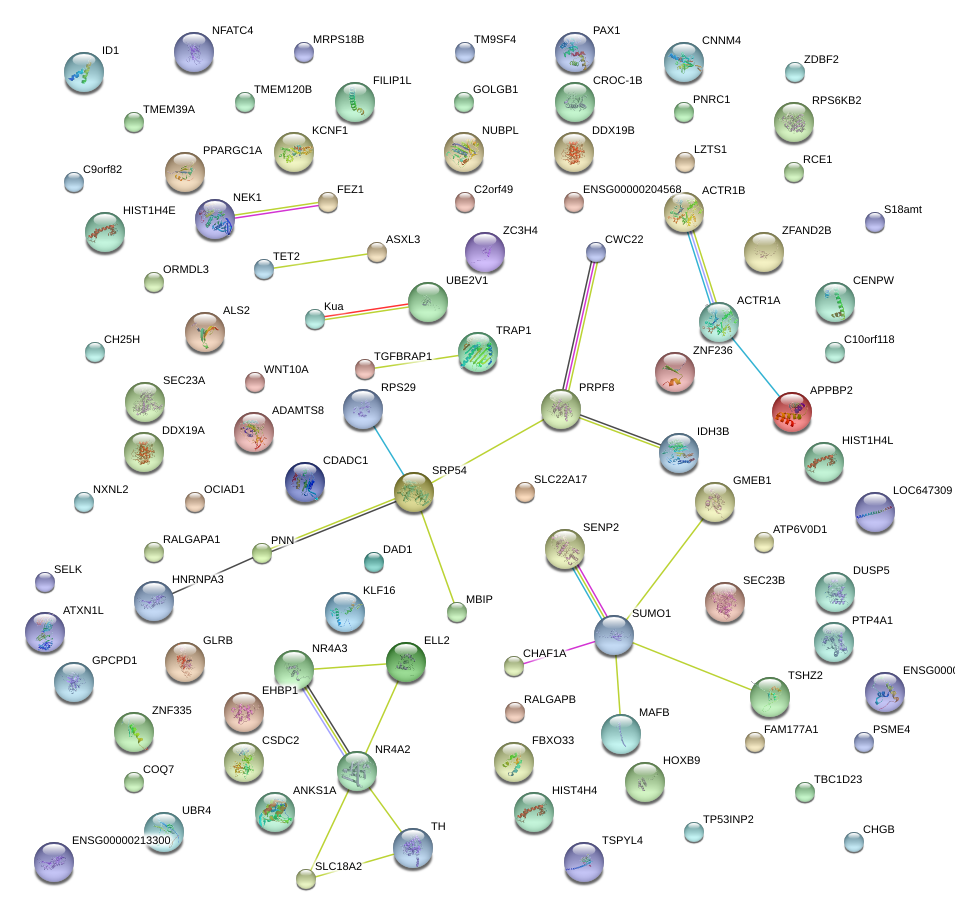
|
chr2_-_156897286
|
5.028
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr3_+_101462375
|
3.352
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr6_+_89848229
|
3.048
|
|
PNRC1
|
proline-rich nuclear receptor coactivator 1
|
|
chr2_+_177785667
|
2.668
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr2_+_219453487
|
2.629
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr2_+_206847658
|
2.619
|
NM_020923
|
ZDBF2
|
zinc finger, DBF-type containing 2
|
|
chr3_-_120665113
|
2.539
|
NM_018266
|
TMEM39A
|
transmembrane protein 39A
|
|
chr14_-_35348086
|
2.464
|
NM_014990
NM_194301
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chr4_-_170770111
|
2.437
|
NM_012224
|
NEK1
|
NIMA (never in mitosis gene a)-related kinase 1
|
|
chr20_+_32755778
|
2.351
|
NM_021202
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr2_+_105320440
|
2.341
|
NM_024093
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr22_+_40286957
|
2.340
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr14_-_35859545
|
2.177
|
NM_001144891
NM_016586
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr18_+_29412538
|
2.160
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr16_+_68938264
|
2.133
|
NM_018332
|
DDX19A
|
DEAD (Asp-Glu-Ala-As) box polypeptide 19A
|
|
chr17_-_55958274
|
2.119
|
NM_006380
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr2_+_105320247
|
2.080
|
|
C2orf49
|
chromosome 2 open reading frame 49
|
|
chr16_-_66072477
|
2.005
|
NM_004691
|
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1
|
|
chr3_-_122951200
|
1.985
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chr6_+_26312758
|
1.923
|
NM_003545
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr14_+_31100330
|
1.884
|
NM_025152
|
NUBPL
|
nucleotide binding protein-like
|
|
chr6_+_30693464
|
1.884
|
NM_014046
|
MRPS18B
|
mitochondrial ribosomal protein S18B
|
|
chr17_-_55958073
|
1.857
|
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2
|
|
chr6_+_126702914
|
1.837
|
NM_001012507
|
CENPW
|
centromere protein W
|
|
chr1_+_169721260
|
1.833
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr3_-_192063158
|
1.824
|
NM_001146686
|
GEMC1
|
geminin coiled-coil domain-containing protein 1
|
|
chr14_-_38971370
|
1.800
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr20_-_38751289
|
1.797
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr16_+_18986721
|
1.754
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr6_+_34964999
|
1.748
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr11_+_66952462
|
1.712
|
NM_003952
|
RPS6KB2
|
ribosomal protein S6 kinase, 70kDa, polypeptide 2
|
|
chr20_+_18436167
|
1.710
|
NM_006363
NM_032985
NM_032986
NM_001172746
|
SEC23B
|
Sec23 homolog B (S. cerevisiae)
|
|
chr3_-_53900888
|
1.693
|
NM_021237
|
SELK
|
selenoprotein K
|
|
chr8_+_128875902
|
1.690
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr14_-_38971049
|
1.680
|
|
FBXO33
|
F-box protein 33
|
|
chr1_-_19409328
|
1.677
|
NM_020765
|
UBR4
|
ubiquitin protein ligase E3 component n-recognin 4
|
|
chr12_+_120635021
|
1.663
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr20_+_21634293
|
1.659
|
NM_006192
|
PAX1
|
paired box 1
|
|
chr16_+_70437394
|
1.659
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr20_-_5539487
|
1.636
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr14_+_34585098
|
1.616
|
NM_173607
|
FAM177A1
|
family with sequence similarity 177, member A1
|
|
chr10_-_115923874
|
1.613
|
|
C10orf118
|
chromosome 10 open reading frame 118
|
|
chr4_+_158216631
|
1.612
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr20_+_29656744
|
1.606
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr14_+_34521945
|
1.603
|
|
SRP54
|
signal recognition particle 54kDa
|
|
chr19_-_52308848
|
1.598
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr4_+_48527798
|
1.589
|
NM_001079840
NM_001079841
NM_017830
NM_001079839
NM_001079842
NM_001168254
|
OCIAD1
|
OCIA domain containing 1
|
|
chr17_-_35337288
|
1.560
|
NM_139280
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr14_-_49122813
|
1.553
|
NM_001030001
NM_001032
|
RPS29
|
ribosomal protein S29
|
|
chr2_-_180580024
|
1.541
|
NM_020943
|
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chr20_-_48163071
|
1.539
|
NM_001032288
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr16_+_18986459
|
1.535
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr14_+_23905984
|
1.511
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr10_+_112247614
|
1.494
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr20_+_5839973
|
1.492
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr14_+_34521854
|
1.480
|
NM_001146282
NM_003136
|
SRP54
|
signal recognition particle 54kDa
|
|
chr2_-_202353898
|
1.475
|
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile)
|
|
chr20_+_36534871
|
1.473
|
NM_020336
|
RALGAPB
|
Ral GTPase activating protein, beta subunit (non-catalytic)
|
|
chr20_-_2592777
|
1.456
|
NM_006899
NM_174855
NM_174856
|
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta
|
|
chr14_-_35347791
|
1.444
|
|
RALGAPA1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic)
|
|
chr17_-_44058612
|
1.430
|
NM_024017
|
HOXB9
|
homeobox B9
|
|
chr2_+_62787534
|
1.429
|
NM_001142614
|
EHBP1
|
EH domain binding protein 1
|
|
chr10_+_118990573
|
1.415
|
NM_003054
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr4_-_23500706
|
1.411
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr2_+_96790298
|
1.407
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr6_-_116681863
|
1.407
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr2_-_234317362
|
1.377
|
NM_001001394
|
DNAJB3
|
DnaJ (Hsp40) homolog, subfamily B, member 3
|
|
chr20_-_44034346
|
1.353
|
|
ZNF335
|
zinc finger protein 335
|
|
chr4_+_106287391
|
1.352
|
NM_001127208
NM_017628
|
TET2
|
tet oncogene family member 2
|
|
chr14_-_38642029
|
1.342
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr11_-_2149591
|
1.340
|
NM_000360
NM_199292
NM_199293
|
TH
|
tyrosine hydroxylase
|
|
chr5_-_95323353
|
1.329
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr9_+_101623957
|
1.313
|
NM_006981
NM_173199
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr13_+_48720047
|
1.309
|
NM_001193478
NM_030911
|
CDADC1
|
cytidine and dCMP deaminase domain containing 1
|
|
chr14_-_38642165
|
1.306
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr6_+_64339869
|
1.299
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr10_-_90957028
|
1.256
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr4_-_187884721
|
1.255
|
|
|
|
|
chr18_-_29412148
|
1.244
|
|
|
|
|
chr2_-_54051290
|
1.242
|
NM_014614
|
PSME4
|
proteasome (prosome, macropain) activator subunit 4
|
|
chr2_+_10969513
|
1.240
|
NM_002236
|
KCNF1
|
potassium voltage-gated channel, subfamily F, member 1
|
|
chr16_-_3707450
|
1.224
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr11_-_66367279
|
1.218
|
|
|
|
|
chr14_-_22127954
|
1.217
|
NM_001344
|
DAD1
|
defender against cell death 1
|
|
chr14_-_22127902
|
1.211
|
|
DAD1
|
defender against cell death 1
|
|
chr16_+_18986417
|
1.196
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr3_-_101077605
|
1.196
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr5_-_95323180
|
1.192
|
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr2_+_219779737
|
1.187
|
NM_138802
|
ZFAND2B
|
zinc finger, AN1-type domain 2B
|
|
chr14_-_22891865
|
1.184
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr20_+_51023159
|
1.182
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr9_-_26882713
|
1.178
|
NM_001167575
NM_024828
|
C9orf82
|
chromosome 9 open reading frame 82
|
|
chr11_-_124871258
|
1.173
|
NM_005103
NM_022549
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I)
|
|
chr14_+_38714111
|
1.171
|
NM_002687
|
PNN
|
pinin, desmosome associated protein
|
|
chr20_+_30161173
|
1.168
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr2_-_156894927
|
1.166
|
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr3_+_186786655
|
1.142
|
NM_021627
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2
|
|
chr20_+_30160952
|
1.141
|
NM_014742
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr11_+_66367458
|
1.137
|
NM_001032279
NM_005133
|
RCE1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae)
|
|
chr19_+_4353658
|
1.135
|
NM_005483
|
CHAF1A
|
chromatin assembly factor 1, subunit A (p150)
|
|
chr2_-_97646758
|
1.125
|
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast)
|
|
chr2_+_108432097
|
1.125
|
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chr20_+_18436525
|
1.122
|
|
SEC23B
|
Sec23 homolog B (S. cerevisiae)
|
|
chr2_+_190014432
|
1.122
|
|
WDR75
|
WD repeat domain 75
|
|
chr2_-_213111525
|
1.121
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr20_-_34835546
|
1.121
|
NM_001145316
NM_024918
NM_001145315
NM_001145317
NM_001145318
|
DSN1
|
DSN1, MIND kinetochore complex component, homolog (S. cerevisiae)
|
|
chr2_-_202811459
|
1.119
|
NM_001005781
NM_001005782
NM_003352
|
SUMO1
|
SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae)
|
|
chr17_+_25828546
|
1.117
|
NM_001007024
NM_001007025
NM_004871
|
GOSR1
|
golgi SNAP receptor complex member 1
|
|
chr1_+_78243181
|
1.116
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr9_-_68552337
|
1.114
|
NM_001085457
|
CBWD5
CBWD3
CBWD1
CBWD6
|
COBW domain containing 5
COBW domain containing 3
COBW domain containing 1
COBW domain containing 6
|
|
chr20_-_44034194
|
1.101
|
NM_022095
|
ZNF335
|
zinc finger protein 335
|
|
chr2_+_108769572
|
1.100
|
NM_144978
|
CCDC138
|
coiled-coil domain containing 138
|
|
chr5_+_50714996
|
1.100
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr15_+_48503860
|
1.099
|
NM_001128610
NM_001128611
NM_005154
|
USP8
|
ubiquitin specific peptidase 8
|
|
chr6_-_128883125
|
1.098
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr17_-_632135
|
1.087
|
NM_016080
|
GLOD4
|
glyoxalase domain containing 4
|
|
chr6_+_10803141
|
1.084
|
NM_017906
|
PAK1IP1
|
PAK1 interacting protein 1
|
|
chr3_-_16281287
|
1.075
|
NM_001047434
NM_206831
|
DPH3
|
DPH3, KTI11 homolog (S. cerevisiae)
|
|
chr2_+_8739563
|
1.074
|
NM_002166
|
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein
|
|
chr9_+_17568952
|
1.061
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr5_+_50714714
|
1.056
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr5_-_50714836
|
1.055
|
|
LOC642366
|
hypothetical LOC642366
|
|
chr7_-_19715127
|
1.055
|
NM_001002926
|
TWISTNB
|
TWIST neighbor
|
|
chr20_+_36810510
|
1.047
|
NM_024855
|
ACTR5
|
ARP5 actin-related protein 5 homolog (yeast)
|
|
chr2_+_183289205
|
1.045
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr5_-_72780107
|
1.041
|
NM_004472
|
FOXD1
|
forkhead box D1
|
|
chr17_+_33761234
|
1.035
|
NM_014598
|
SOCS7
|
suppressor of cytokine signaling 7
|
|
chr5_-_89861048
|
1.029
|
NM_198273
|
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3
|
|
chr9_-_168913
|
1.027
|
NM_001145355
NM_001145356
NM_018491
|
CBWD2
CBWD1
|
COBW domain containing 2
COBW domain containing 1
|
|
chr14_+_37748031
|
1.025
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr12_+_105692466
|
1.024
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr2_-_202353818
|
1.022
|
|
|
|
|
chr6_-_3102690
|
1.020
|
NM_001069
|
TUBB2A
|
tubulin, beta 2A
|
|
chr9_-_32540818
|
1.019
|
|
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase
|
|
chr17_+_55051818
|
1.018
|
NM_004859
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr2_+_33025872
|
1.012
|
NM_206943
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr6_+_14033076
|
1.010
|
NM_001165032
NM_152737
|
RNF182
|
ring finger protein 182
|
|
chr6_+_26705110
|
1.008
|
NM_013375
|
ABT1
|
activator of basal transcription 1
|
|
chr6_+_56927882
|
1.008
|
|
BEND6
|
BEN domain containing 6
|
|
chr10_+_103901922
|
1.003
|
NM_004741
|
NOLC1
|
nucleolar and coiled-body phosphoprotein 1
|
|
chr18_-_39111243
|
1.002
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr11_-_76862197
|
0.999
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr9_+_4669557
|
0.997
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr4_+_72271218
|
0.995
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr5_-_133332173
|
0.995
|
NM_020199
|
C5orf15
|
chromosome 5 open reading frame 15
|
|
chr2_+_95432150
|
0.994
|
NM_016044
|
FAHD2A
|
fumarylacetoacetate hydrolase domain containing 2A
|
|
chr2_+_190014403
|
0.988
|
NM_032168
|
WDR75
|
WD repeat domain 75
|
|
chr12_+_61147083
|
0.982
|
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr2_-_25047988
|
0.981
|
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr6_+_106653429
|
0.980
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr6_+_139497933
|
0.978
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr8_+_64243741
|
0.971
|
|
YTHDF3
|
YTH domain family, member 3
|
|
chr17_+_632262
|
0.970
|
NM_018146
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr1_-_58938323
|
0.968
|
NM_001085487
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1
|
|
chr4_-_56948345
|
0.959
|
NM_181806
|
AASDH
|
aminoadipate-semialdehyde dehydrogenase
|
|
chr2_-_60634271
|
0.959
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr9_+_115023628
|
0.957
|
NM_001859
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr14_-_23781607
|
0.957
|
NM_001099274
NM_012461
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2
|
|
chr3_+_142143329
|
0.955
|
NM_001104647
NM_018155
|
SLC25A36
|
solute carrier family 25, member 36
|
|
chr2_-_97646970
|
0.954
|
NM_005735
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast)
|
|
chr2_+_30307900
|
0.953
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr3_+_171167243
|
0.949
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr9_+_115023684
|
0.947
|
|
SLC31A1
|
solute carrier family 31 (copper transporters), member 1
|
|
chr5_+_61744322
|
0.945
|
NM_016338
|
IPO11
|
importin 11
|
|
chr14_+_73423260
|
0.944
|
NM_021188
|
ZNF410
|
zinc finger protein 410
|
|
chr15_+_32304477
|
0.941
|
NM_016454
|
TMEM85
|
transmembrane protein 85
|
|
chr4_-_186584031
|
0.938
|
NM_018359
|
UFSP2
|
UFM1-specific peptidase 2
|
|
chr6_-_53321658
|
0.936
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr12_-_10657397
|
0.932
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr2_-_43676603
|
0.929
|
NM_001083953
NM_022065
|
THADA
|
thyroid adenoma associated
|
|
chr7_-_25131427
|
0.928
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr5_-_64813459
|
0.925
|
NM_197941
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6
|
|
chr6_+_30137014
|
0.923
|
NM_014596
NM_170783
|
ZNRD1
|
zinc ribbon domain containing 1
|
|
chr2_+_170149348
|
0.922
|
|
PPIG
|
peptidylprolyl isomerase G (cyclophilin G)
|
|
chr2_-_179051471
|
0.917
|
NM_001135212
NM_181342
|
FKBP7
|
FK506 binding protein 7
|
|
chr6_-_53321545
|
0.917
|
|
ELOVL5
|
ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3-like, yeast)
|
|
chr13_+_21143083
|
0.916
|
NM_002010
|
FGF9
|
fibroblast growth factor 9 (glia-activating factor)
|
|
chr2_-_10747553
|
0.915
|
NM_024894
|
NOL10
|
nucleolar protein 10
|
|
chr20_-_39379533
|
0.911
|
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr17_+_24095135
|
0.902
|
NM_004295
|
TRAF4
|
TNF receptor-associated factor 4
|
|
chr3_+_114733907
|
0.902
|
NM_017699
|
SIDT1
|
SID1 transmembrane family, member 1
|
|
chr22_-_25209761
|
0.900
|
NM_022081
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr2_-_10747438
|
0.899
|
|
NOL10
|
nucleolar protein 10
|
|
chr4_-_120595218
|
0.896
|
|
|
|
|
chr7_-_25131386
|
0.894
|
|
CYCS
|
cytochrome c, somatic
|
|
chr9_+_124066973
|
0.893
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr6_+_114285209
|
0.893
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr17_-_38229790
|
0.889
|
NM_003766
|
BECN1
|
beclin 1, autophagy related
|
|
chr9_-_72218098
|
0.888
|
|
KLF9
|
Kruppel-like factor 9
|
|
chr14_+_103165244
|
0.887
|
NM_001130107
NM_005552
NM_182923
|
KLC1
|
kinesin light chain 1
|
|
chr13_-_79810972
|
0.880
|
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr6_+_116798791
|
0.879
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr22_-_42589543
|
0.877
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr20_-_10602429
|
0.876
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr15_+_36533595
|
0.875
|
NM_001042429
NM_173611
|
FAM98B
|
family with sequence similarity 98, member B
|
|
chr2_+_113911737
|
0.874
|
NM_172003
|
CBWD2
CBWD1
|
COBW domain containing 2
COBW domain containing 1
|
|
chr12_-_62902279
|
0.874
|
NM_152440
|
C12orf66
|
chromosome 12 open reading frame 66
|
|
chr12_+_58276114
|
0.871
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr17_-_46553083
|
0.869
|
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr10_-_45410197
|
0.869
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|