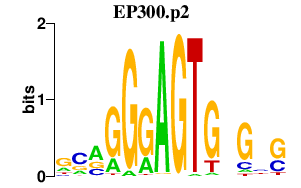
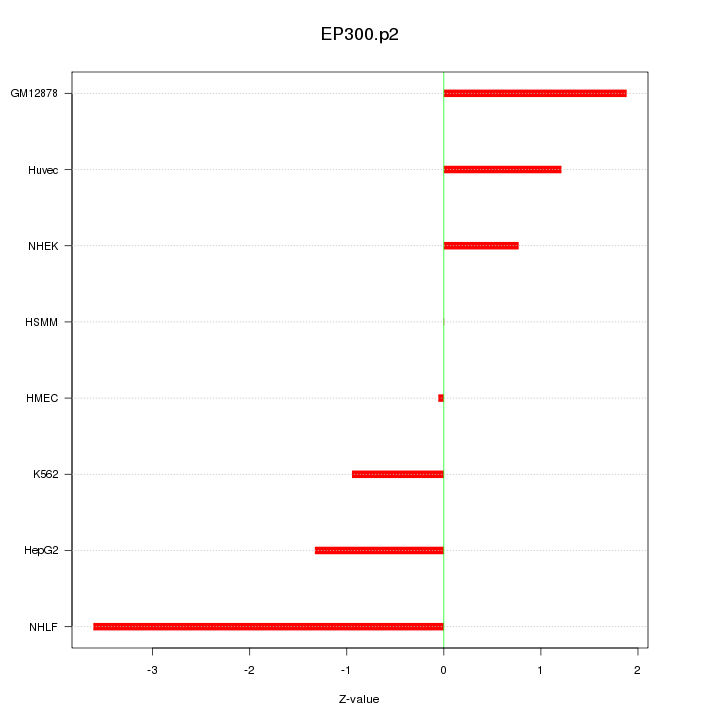
|
chr12_+_6808025
|
1.750
|
|
LEPREL2
|
leprecan-like 2
|
|
chr13_-_43259032
|
1.727
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr4_-_122213018
|
1.599
|
NM_024574
|
C4orf31
|
chromosome 4 open reading frame 31
|
|
chr1_-_29322993
|
1.562
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chr4_+_172971149
|
1.538
|
NM_001034845
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6
|
|
chr14_-_50480857
|
1.497
|
NM_001163940
NM_002863
|
PYGL
|
phosphorylase, glycogen, liver
|
|
chr9_-_129729518
|
1.400
|
NM_173492
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1
|
|
chr9_+_70107602
|
1.328
|
NM_199135
|
FOXD4L3
FOXD4L6
|
forkhead box D4-like 3
forkhead box D4-like 6
|
|
chr17_+_56843893
|
1.299
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr17_+_69939130
|
1.270
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr20_+_43468046
|
1.248
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr7_-_117300763
|
1.162
|
NM_033427
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr7_+_128257698
|
1.129
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr9_+_114552871
|
1.118
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr2_-_230287442
|
1.117
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr22_-_27405708
|
1.087
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr19_+_12966970
|
1.085
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr16_-_85099941
|
1.083
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr4_+_183302105
|
1.053
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr10_-_134449467
|
1.030
|
NM_177400
|
NKX6-2
|
NK6 homeobox 2
|
|
chr19_+_41039649
|
1.027
|
NM_032123
NM_199179
NM_199180
|
KIRREL2
|
kin of IRRE like 2 (Drosophila)
|
|
chr21_+_44941514
|
1.023
|
NM_198699
|
KRTAP10-12
|
keratin associated protein 10-12
|
|
chr19_-_1352474
|
1.018
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr8_-_23077392
|
0.996
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr2_-_200030944
|
0.975
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr6_+_43151922
|
0.973
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr1_-_154913712
|
0.967
|
NM_006617
|
NES
|
nestin
|
|
chr3_-_127558747
|
0.958
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr20_+_43468617
|
0.955
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chrX_+_37093448
|
0.953
|
NM_000950
NM_001142395
NM_001173486
NM_001173489
NM_001173490
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1
|
|
chr1_-_12599934
|
0.938
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr15_-_35177664
|
0.927
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr1_+_179148935
|
0.918
|
NM_020950
|
KIAA1614
|
KIAA1614
|
|
chr1_-_245342150
|
0.913
|
NM_207401
|
C1orf229
|
chromosome 1 open reading frame 229
|
|
chr6_+_99389300
|
0.903
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr3_+_160964574
|
0.903
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr17_-_72045218
|
0.880
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr12_-_26169074
|
0.872
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr17_-_76985538
|
0.835
|
|
|
|
|
chr2_-_204108151
|
0.822
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr1_+_56883333
|
0.813
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr8_-_134378630
|
0.776
|
NM_001135242
NM_006096
|
NDRG1
|
N-myc downstream regulated 1
|
|
chr12_+_6179736
|
0.768
|
NM_001769
|
CD9
|
CD9 molecule
|
|
chr14_-_60260211
|
0.765
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr12_-_49853155
|
0.755
|
NM_001173452
NM_001173453
NM_005653
|
TFCP2
|
transcription factor CP2
|
|
chr14_+_92049396
|
0.752
|
NM_024832
|
RIN3
|
Ras and Rab interactor 3
|
|
chr2_+_191818124
|
0.735
|
NM_001130158
NM_012223
NM_001161819
|
MYO1B
|
myosin IB
|
|
chr8_-_111056099
|
0.732
|
NM_014379
|
KCNV1
|
potassium channel, subfamily V, member 1
|
|
chr9_+_2612052
|
0.731
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr20_-_61755145
|
0.730
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr15_+_90738108
|
0.725
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr3_+_35656012
|
0.723
|
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr11_-_103540233
|
0.717
|
NM_025208
NM_033135
|
PDGFD
|
platelet derived growth factor D
|
|
chr8_+_106400322
|
0.716
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr21_+_37300732
|
0.715
|
NM_018962
|
DSCR6
|
Down syndrome critical region gene 6
|
|
chrX_+_71047344
|
0.710
|
NM_001013627
|
NHSL2
|
NHS-like 2
|
|
chr4_-_159863889
|
0.694
|
NM_005038
|
PPID
|
peptidylprolyl isomerase D
|
|
chr12_-_56445653
|
0.688
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr11_+_131286513
|
0.688
|
|
NTM
|
neurotrimin
|
|
chr11_+_104986009
|
0.686
|
NM_000829
NM_001077243
NM_001077244
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr19_-_51608665
|
0.674
|
NM_032040
|
CCDC8
|
coiled-coil domain containing 8
|
|
chr2_+_20729904
|
0.672
|
NM_182828
|
GDF7
|
growth differentiation factor 7
|
|
chr3_-_130807865
|
0.671
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr18_+_32021418
|
0.667
|
NM_017947
|
MOCOS
|
molybdenum cofactor sulfurase
|
|
chr22_+_39404940
|
0.664
|
NM_005297
|
MCHR1
|
melanin-concentrating hormone receptor 1
|
|
chr14_-_87859284
|
0.662
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr17_+_76988134
|
0.655
|
NM_001080519
|
BAHCC1
|
BAH domain and coiled-coil containing 1
|
|
chr1_-_94779727
|
0.650
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr20_-_26137755
|
0.648
|
|
LOC284801
|
hypothetical protein LOC284801
|
|
chr13_-_37341859
|
0.644
|
NM_001135955
NM_001135956
NM_001135957
NM_001135958
NM_003306
NM_016179
|
TRPC4
|
transient receptor potential cation channel, subfamily C, member 4
|
|
chr8_-_122722674
|
0.630
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr9_-_108416
|
0.629
|
NM_207305
|
FOXD4
|
forkhead box D4
|
|
chr19_-_60573552
|
0.629
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr11_+_104986620
|
0.628
|
NM_001112812
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr5_-_139992621
|
0.627
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr20_-_61108831
|
0.625
|
NM_080606
|
BHLHE23
|
basic helix-loop-helix family, member e23
|
|
chr18_+_53253775
|
0.617
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr9_-_113977285
|
0.616
|
NM_022486
|
SUSD1
|
sushi domain containing 1
|
|
chr16_-_10183616
|
0.613
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr11_+_696607
|
0.612
|
|
EPS8L2
|
EPS8-like 2
|
|
chr15_-_80123935
|
0.610
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr10_+_52504239
|
0.608
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr5_-_131658838
|
0.608
|
|
|
|
|
chr1_+_154878344
|
0.599
|
NM_021948
NM_198427
|
BCAN
|
brevican
|
|
chr6_-_72187168
|
0.597
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr1_+_41022066
|
0.597
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr3_+_156280539
|
0.593
|
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_-_186353421
|
0.589
|
NM_001025266
|
C3orf70
|
chromosome 3 open reading frame 70
|
|
chr9_-_116920232
|
0.587
|
NM_002160
|
TNC
|
tenascin C
|
|
chr9_-_132804057
|
0.586
|
NM_032843
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr11_-_103539895
|
0.584
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr20_+_34143983
|
0.579
|
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr11_+_129823667
|
0.576
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr5_+_82803248
|
0.576
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr15_+_97010219
|
0.574
|
NM_000875
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr14_+_101297887
|
0.572
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr14_+_91859904
|
0.569
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr2_+_46378405
|
0.567
|
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr20_-_4177519
|
0.567
|
NM_000678
|
ADRA1D
|
adrenergic, alpha-1D-, receptor
|
|
chr5_-_114533448
|
0.563
|
|
TRIM36
|
tripartite motif containing 36
|
|
chr10_-_125843092
|
0.562
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr14_+_55654607
|
0.560
|
NM_021255
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr5_+_42459568
|
0.560
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr11_-_103540275
|
0.558
|
|
PDGFD
|
platelet derived growth factor D
|
|
chr12_-_113328271
|
0.557
|
NM_181486
|
TBX5
|
T-box 5
|
|
chr17_-_1874927
|
0.556
|
NM_178568
|
RTN4RL1
|
reticulon 4 receptor-like 1
|
|
chr11_+_68208583
|
0.555
|
|
GAL
|
galanin prepropeptide
|
|
chr8_+_22465368
|
0.554
|
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr1_+_202308814
|
0.552
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr11_-_122571198
|
0.551
|
NM_024769
|
CLMP
|
CXADR-like membrane protein
|
|
chr3_+_112273352
|
0.550
|
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr14_-_60260780
|
0.546
|
|
SIX4
|
SIX homeobox 4
|
|
chr2_+_109112428
|
0.545
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr9_+_86473273
|
0.545
|
NM_001018064
NM_006180
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr20_+_55569542
|
0.545
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr20_+_3000838
|
0.542
|
|
OXT
|
oxytocin, prepropeptide
|
|
chr17_+_14145051
|
0.542
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr11_-_64267037
|
0.538
|
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr18_-_26935427
|
0.536
|
|
DSC2
|
desmocollin 2
|
|
chr6_+_73388713
|
0.534
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr15_+_66658626
|
0.532
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr19_+_447453
|
0.531
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr19_+_34709330
|
0.528
|
NM_001146339
|
VSTM2B
|
V-set and transmembrane domain containing 2B
|
|
chr1_-_225572419
|
0.526
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr5_+_96024434
|
0.522
|
|
CAST
|
calpastatin
|
|
chr2_+_130703384
|
0.521
|
|
|
|
|
chr13_-_60887210
|
0.518
|
|
PCDH20
|
protocadherin 20
|
|
chr11_+_68208554
|
0.518
|
NM_015973
|
GAL
|
galanin prepropeptide
|
|
chrX_+_1347692
|
0.514
|
NM_001161529
NM_001161531
NM_001161532
NM_006140
NM_172245
NM_172246
NM_172249
|
CSF2RA
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr19_+_16157198
|
0.514
|
NM_014077
|
FAM32A
|
family with sequence similarity 32, member A
|
|
chr6_+_101948102
|
0.513
|
|
|
|
|
chr19_-_15204767
|
0.513
|
NM_001142886
|
EPHX3
|
epoxide hydrolase 3
|
|
chr1_+_219119358
|
0.513
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr1_+_213323182
|
0.509
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr1_+_154351200
|
0.509
|
|
LMNA
|
lamin A/C
|
|
chr2_+_200031333
|
0.507
|
|
|
|
|
chr1_-_182273114
|
0.507
|
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr12_+_52665196
|
0.505
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr18_-_26935883
|
0.504
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr1_+_155130146
|
0.502
|
NM_001080471
|
PEAR1
|
platelet endothelial aggregation receptor 1
|
|
chr5_+_128823904
|
0.501
|
NM_133638
|
ADAMTS19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19
|
|
chr8_+_21971317
|
0.500
|
NM_001114135
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr14_-_106105805
|
0.500
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr9_-_132804243
|
0.498
|
NM_001145106
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr16_+_2461500
|
0.494
|
NM_006181
|
NTN3
|
netrin 3
|
|
chr18_+_65219114
|
0.483
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chrX_-_99551926
|
0.483
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr17_+_40089270
|
0.483
|
NM_001145080
|
C17orf104
|
chromosome 17 open reading frame 104
|
|
chr12_-_113606322
|
0.482
|
NM_005996
NM_016569
|
TBX3
|
T-box 3
|
|
chr20_-_22512893
|
0.481
|
|
FOXA2
|
forkhead box A2
|
|
chr12_-_53153639
|
0.480
|
NM_144594
|
GTSF1
|
gametocyte specific factor 1
|
|
chr5_-_92942680
|
0.477
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr11_+_10428443
|
0.476
|
NM_001172431
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr10_-_103991219
|
0.475
|
NM_005029
|
PITX3
|
paired-like homeodomain 3
|
|
chr6_+_73388291
|
0.474
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr11_-_64266916
|
0.473
|
|
|
|
|
chr19_-_40325194
|
0.473
|
|
LGI4
|
leucine-rich repeat LGI family, member 4
|
|
chr4_+_156900053
|
0.472
|
|
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3
|
|
chr22_+_49386044
|
0.468
|
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr1_-_176273680
|
0.467
|
|
LOC730102
|
quinone oxidoreductase-like protein 2 pseudogene
|
|
chr2_-_200028977
|
0.467
|
|
SATB2
|
SATB homeobox 2
|
|
chr10_-_126839546
|
0.465
|
NM_001083914
|
CTBP2
|
C-terminal binding protein 2
|
|
chr19_-_11390948
|
0.465
|
NM_001035223
NM_001161616
|
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3
|
|
chr12_+_54760153
|
0.465
|
NM_001005915
NM_001982
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr14_+_55654900
|
0.464
|
|
PELI2
|
pellino homolog 2 (Drosophila)
|
|
chr10_+_125415860
|
0.464
|
NM_153442
|
GPR26
|
G protein-coupled receptor 26
|
|
chr15_+_72695248
|
0.462
|
|
CLK3
|
CDC-like kinase 3
|
|
chr22_+_36286350
|
0.461
|
NM_152243
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1
|
|
chr7_-_150605112
|
0.460
|
NM_003078
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3
|
|
chr8_+_22465171
|
0.458
|
NM_005775
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr17_+_8865547
|
0.457
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr5_+_119827903
|
0.453
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr3_+_19164973
|
0.451
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr4_-_86106567
|
0.451
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr1_-_13712744
|
0.448
|
NM_001010847
|
LRRC38
|
leucine rich repeat containing 38
|
|
chr16_+_85158357
|
0.448
|
NM_005251
|
FOXC2
|
forkhead box C2 (MFH-1, mesenchyme forkhead 1)
|
|
chr3_-_140148671
|
0.447
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr19_+_50693570
|
0.447
|
NM_001080401
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative)
|
|
chr1_+_219119430
|
0.446
|
|
HLX
|
H2.0-like homeobox
|
|
chr5_+_119827797
|
0.445
|
|
PRR16
|
proline rich 16
|
|
chr12_+_6363482
|
0.445
|
NM_002342
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3)
|
|
chr7_+_142262669
|
0.445
|
NM_004445
|
EPHB6
|
EPH receptor B6
|
|
chr12_+_117903682
|
0.444
|
NM_194286
|
SRRM4
|
serine/arginine repetitive matrix 4
|
|
chr11_-_27678551
|
0.443
|
NM_001143813
NM_001143814
NM_001709
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_+_67965517
|
0.440
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr2_+_113973130
|
0.440
|
NM_012184
|
FOXD4L1
|
forkhead box D4-like 1
|
|
chr6_+_118335381
|
0.440
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr6_+_1335067
|
0.435
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr2_+_182030173
|
0.434
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr8_+_22513043
|
0.434
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr3_+_73019858
|
0.433
|
NM_001080393
|
GXYLT2
|
glucoside xylosyltransferase 2
|
|
chr19_-_18749728
|
0.432
|
|
|
|
|
chr1_+_156229934
|
0.432
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr11_-_72111027
|
0.431
|
NM_001135190
NM_015242
|
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr12_+_55896844
|
0.429
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr15_-_80125514
|
0.428
|
|
MEX3B
|
mex-3 homolog B (C. elegans)
|
|
chr10_+_102782156
|
0.427
|
|
SFXN3
|
sideroflexin 3
|
|
chrX_-_78509367
|
0.427
|
|
ITM2A
|
integral membrane protein 2A
|
|
chr9_-_131444098
|
0.423
|
NM_017873
NM_177999
|
ASB6
|
ankyrin repeat and SOCS box containing 6
|
|
chr6_+_30082331
|
0.423
|
|
HLA-J
|
major histocompatibility complex, class I, J (pseudogene)
|