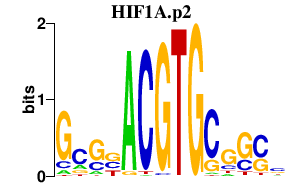
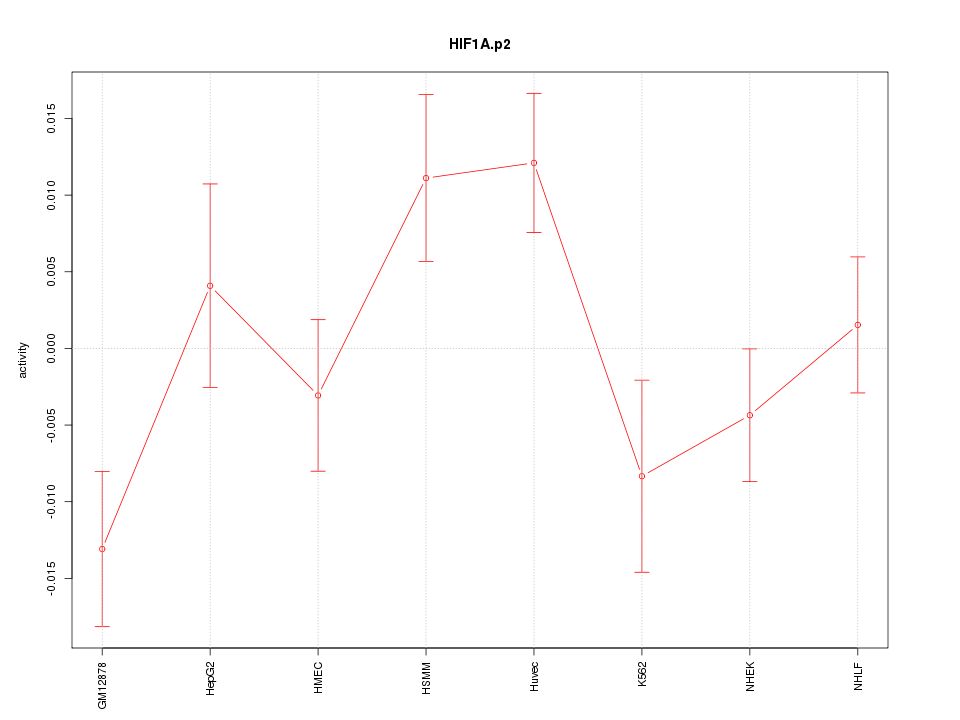
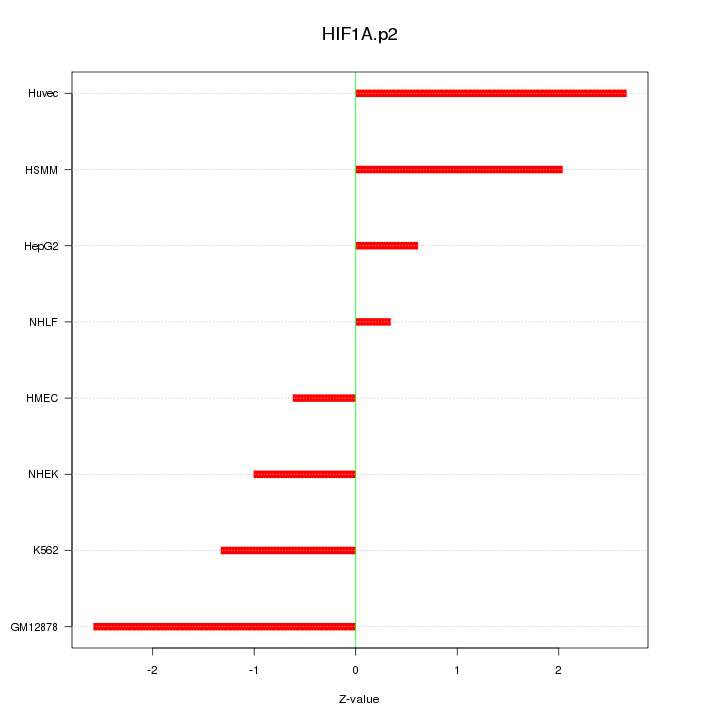
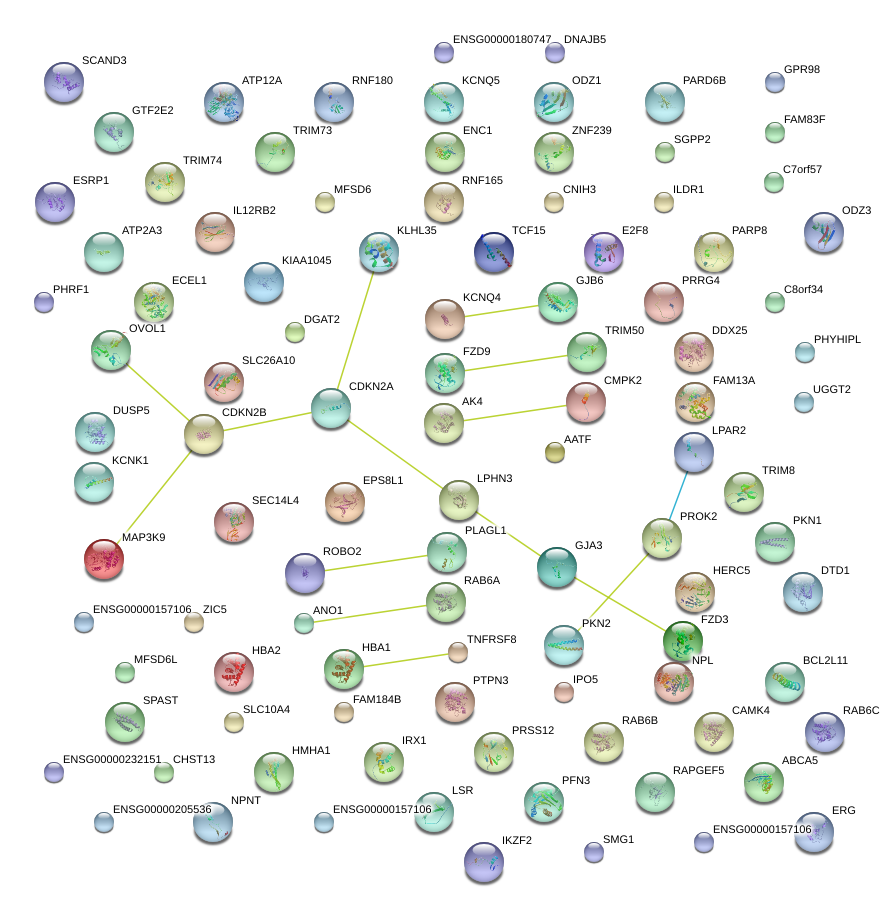
|
chr1_+_12046020
|
1.229
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr10_-_60606195
|
1.176
|
|
|
|
|
chr11_+_32808045
|
1.055
|
NM_024081
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr5_-_176760242
|
1.025
|
NM_001029886
|
PFN3
|
profilin 3
|
|
chr3_-_71916877
|
1.002
|
NM_001126128
NM_021935
|
PROK2
|
prokineticin 2
|
|
chr6_-_144427340
|
0.961
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr2_+_222997456
|
0.919
|
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr1_+_181025494
|
0.894
|
|
NPL
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
|
|
chr20_+_48781457
|
0.888
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr4_+_107036042
|
0.886
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr17_+_32380287
|
0.873
|
NM_012138
|
AATF
|
apoptosis antagonizing transcription factor
|
|
chr2_+_222997529
|
0.856
|
NM_152386
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr16_+_166678
|
0.799
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr2_+_32142083
|
0.793
|
NM_014946
NM_199436
|
SPAST
|
spastin
|
|
chr17_-_3814334
|
0.778
|
NM_005173
NM_174953
NM_174954
NM_174955
NM_174956
NM_174957
NM_174958
|
ATP2A3
|
ATPase, Ca++ transporting, ubiquitous
|
|
chr22_+_38720881
|
0.776
|
NM_138435
|
FAM83F
|
family with sequence similarity 83, member F
|
|
chr2_-_213724577
|
0.758
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr2_+_190981257
|
0.753
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr3_-_123223658
|
0.724
|
NM_175924
|
ILDR1
|
immunoglobulin-like domain containing receptor 1
|
|
chr4_+_107036076
|
0.710
|
|
NPNT
|
nephronectin
|
|
chr1_+_67546143
|
0.702
|
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chr11_+_69602055
|
0.679
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr16_+_162845
|
0.664
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr4_+_89597257
|
0.663
|
NM_016323
|
HERC5
|
hect domain and RLD 5
|
|
chr13_-_99422163
|
0.658
|
NM_033132
|
ZIC5
|
Zic family member 5 (odd-paired homolog, Drosophila)
|
|
chr20_-_538909
|
0.643
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr1_+_231816372
|
0.632
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr3_+_127743868
|
0.625
|
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr1_-_146543102
|
0.620
|
|
|
|
|
chr18_+_42167904
|
0.595
|
|
RNF165
|
ring finger protein 165
|
|
chr3_+_77171938
|
0.592
|
NM_002942
|
ROBO2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila)
|
|
chr6_+_73388713
|
0.589
|
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr17_-_64834780
|
0.586
|
NM_172232
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5
|
|
chr6_-_28663045
|
0.581
|
NM_052923
|
SCAND3
|
SCAN domain containing 3
|
|
chr13_-_95503566
|
0.580
|
NM_020121
|
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2
|
|
chr4_-_90251571
|
0.573
|
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr1_+_222870573
|
0.573
|
|
CNIH3
|
cornichon homolog 3 (Drosophila)
|
|
chr19_+_1022208
|
0.565
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr10_+_60606352
|
0.562
|
NM_032439
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like
|
|
chr12_+_56299959
|
0.553
|
NM_133489
|
SLC26A10
|
solute carrier family 26, member 10
|
|
chr5_+_49998677
|
0.551
|
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr10_+_112247614
|
0.547
|
NM_004419
|
DUSP5
|
dual specificity phosphatase 5
|
|
chr11_+_65311104
|
0.539
|
NM_004561
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr8_+_95722448
|
0.527
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr4_+_183302105
|
0.524
|
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr4_+_48180033
|
0.506
|
NM_152679
|
SLC10A4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4
|
|
chr14_-_70345485
|
0.503
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr4_-_119493322
|
0.494
|
NM_003619
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin)
|
|
chr9_+_34948171
|
0.490
|
NM_015297
|
KIAA1045
|
KIAA1045
|
|
chr1_-_142536082
|
0.488
|
|
|
|
|
chr2_-_233060774
|
0.486
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr13_+_24152548
|
0.484
|
NM_001185085
NM_001676
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide
|
|
chr11_+_75157414
|
0.483
|
NM_032564
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr11_-_74819165
|
0.482
|
NM_001039548
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr17_-_8643163
|
0.482
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr8_+_69405510
|
0.481
|
NM_001195639
NM_052958
|
C8orf34
|
chromosome 8 open reading frame 34
|
|
chr21_-_38954433
|
0.480
|
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr13_-_19704533
|
0.480
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr19_+_40431678
|
0.479
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr2_+_111594921
|
0.476
|
NM_006538
NM_138621
NM_207002
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr19_+_60283571
|
0.475
|
NM_017729
|
EPS8L1
|
EPS8-like 1
|
|
chr9_-_111300350
|
0.469
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr19_-_19599973
|
0.465
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr13_+_97426713
|
0.463
|
|
IPO5
|
importin 5
|
|
chr1_+_67545634
|
0.457
|
NM_001559
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chr1_+_65385799
|
0.447
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr7_-_22363036
|
0.444
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr7_+_48041632
|
0.440
|
NM_001100159
|
C7orf57
|
chromosome 7 open reading frame 57
|
|
chr9_-_21965131
|
0.439
|
NM_000077
NM_001195132
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr2_-_6923215
|
0.437
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr11_-_19219720
|
0.436
|
|
E2F8
|
E2F transcription factor 8
|
|
chr1_-_173428455
|
0.436
|
NM_001162895
NM_014656
NM_001162893
NM_001162894
|
KIAA0040
|
KIAA0040
|
|
chr11_+_65311142
|
0.433
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr13_+_78068246
|
0.430
|
|
LOC780529
|
hypothetical LOC780529
|
|
chr5_+_63497408
|
0.429
|
NM_001113561
NM_178532
|
RNF180
|
ring finger protein 180
|
|
chr7_-_72077805
|
0.425
|
NM_198853
|
TRIM73
TRIM74
|
tripartite motif containing 73
tripartite motif containing 74
|
|
chr11_+_125279481
|
0.423
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr1_+_231816540
|
0.417
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr20_+_18516541
|
0.417
|
NM_080820
|
DTD1
|
D-tyrosyl-tRNA deacylase 1 homolog (S. cerevisiae)
|
|
chr10_+_104394185
|
0.414
|
NM_030912
|
TRIM8
|
tripartite motif containing 8
|
|
chr13_-_19633182
|
0.410
|
NM_021954
|
GJA3
|
gap junction protein, alpha 3, 46kDa
|
|
chr11_-_73149705
|
0.409
|
NM_002869
NM_198896
|
RAB6A
|
RAB6A, member RAS oncogene family
|
|
chr5_+_89890366
|
0.408
|
NM_032119
|
GPR98
|
G protein-coupled receptor 98
|
|
chr1_+_88922424
|
0.405
|
NM_006256
|
PKN2
|
protein kinase N2
|
|
chr10_-_43390014
|
0.405
|
NM_001099282
NM_001099283
NM_001099284
|
ZNF239
|
zinc finger protein 239
|
|
chr8_+_28407613
|
0.404
|
NM_017412
|
FZD3
|
frizzled homolog 3 (Drosophila)
|
|
chr16_-_18845223
|
0.401
|
NM_015092
|
SMG1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr4_-_17392232
|
0.399
|
NM_015688
|
FAM184B
|
family with sequence similarity 184, member B
|
|
chr11_+_566482
|
0.398
|
NM_020901
|
PHRF1
|
PHD and ring finger domains 1
|
|
chr1_+_222870798
|
0.393
|
NM_152495
|
CNIH3
|
cornichon homolog 3 (Drosophila)
|
|
chr8_-_30635112
|
0.386
|
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa
|
|
chr5_+_3649167
|
0.384
|
NM_024337
|
IRX1
|
iroquois homeobox 1
|
|
chr5_+_110587546
|
0.379
|
NM_001744
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV
|
|
chr22_-_29231597
|
0.376
|
NM_001161368
NM_174977
|
SEC14L4
|
SEC14-like 4 (S. cerevisiae)
|
|
chr17_-_42924828
|
0.376
|
|
MRPL45P2
|
mitochondrial ribosomal protein L45 pseudogene 2
|
|
chr8_+_75050930
|
0.375
|
NM_001040613
NM_017866
|
TMEM70
|
transmembrane protein 70
|
|
chr1_-_146543016
|
0.373
|
|
|
|
|
chr7_+_26298018
|
0.373
|
NM_013322
|
SNX10
|
sorting nexin 10
|
|
chr11_+_133706977
|
0.371
|
NM_138342
|
GLB1L2
|
galactosidase, beta 1-like 2
|
|
chr16_-_66571863
|
0.371
|
NM_001129758
NM_022357
|
DPEP3
|
dipeptidase 3
|
|
chr1_+_191357906
|
0.363
|
|
CDC73
|
cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr8_+_31616809
|
0.362
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr2_-_96174865
|
0.360
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr10_+_52420936
|
0.358
|
NM_001098512
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr18_+_53253775
|
0.355
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr2_+_17585462
|
0.355
|
|
VSNL1
|
visinin-like 1
|
|
chr19_-_55999677
|
0.353
|
NM_199250
NM_199249
|
C19orf48
|
chromosome 19 open reading frame 48
|
|
chr5_+_176951802
|
0.352
|
NM_017510
|
TMED9
|
transmembrane emp24 protein transport domain containing 9
|
|
chrX_-_131919896
|
0.352
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr2_+_122229590
|
0.350
|
NM_004622
|
TSN
|
translin
|
|
chr8_-_9046460
|
0.350
|
|
PPP1R3B
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B
|
|
chr1_-_180840016
|
0.349
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chr7_+_115637795
|
0.348
|
NM_015641
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr10_-_98470164
|
0.348
|
NM_152309
|
PIK3AP1
|
phosphoinositide-3-kinase adaptor protein 1
|
|
chr8_-_30635207
|
0.347
|
NM_002095
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa
|
|
chr22_+_24290738
|
0.347
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr2_+_17585287
|
0.345
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chr1_+_20385164
|
0.345
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr13_-_40604587
|
0.341
|
NM_152903
|
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6
|
|
chr17_-_34084682
|
0.339
|
NM_001130677
|
C17orf96
|
chromosome 17 open reading frame 96
|
|
chr2_-_60633908
|
0.338
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr12_-_123965099
|
0.338
|
|
UBC
|
ubiquitin C
|
|
chr19_+_2427122
|
0.337
|
NM_015675
|
GADD45B
|
growth arrest and DNA-damage-inducible, beta
|
|
chr11_+_104986620
|
0.336
|
NM_001112812
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr2_-_112729072
|
0.332
|
NM_032494
|
ZC3H8
|
zinc finger CCCH-type containing 8
|
|
chr11_-_65895640
|
0.330
|
NM_001532
|
SLC29A2
|
solute carrier family 29 (nucleoside transporters), member 2
|
|
chr2_-_101370291
|
0.330
|
|
CREG2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr20_+_41519882
|
0.330
|
NM_006275
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr2_+_219432789
|
0.330
|
NM_006522
|
WNT6
|
wingless-type MMTV integration site family, member 6
|
|
chr1_-_146398501
|
0.330
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr9_+_34948320
|
0.329
|
|
KIAA1045
|
KIAA1045
|
|
chr6_+_111515341
|
0.328
|
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr11_+_133707107
|
0.327
|
|
GLB1L2
|
galactosidase, beta 1-like 2
|
|
chr15_-_76700563
|
0.325
|
NM_000743
NM_001166694
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3
|
|
chr12_+_56230073
|
0.323
|
|
KIF5A
|
kinesin family member 5A
|
|
chr17_+_50697355
|
0.320
|
|
HLF
|
hepatic leukemia factor
|
|
chr11_-_565848
|
0.320
|
|
LOC143666
|
hypothetical LOC143666
|
|
chr10_-_95350949
|
0.319
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr17_+_45778381
|
0.318
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr16_+_73739890
|
0.316
|
NM_153688
|
ZFP1
|
zinc finger protein 1 homolog (mouse)
|
|
chr10_+_105334704
|
0.316
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr9_+_129199204
|
0.315
|
NM_014580
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8
|
|
chr6_+_111515473
|
0.313
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr6_+_1336077
|
0.313
|
|
|
|
|
chr6_-_153346283
|
0.312
|
NM_001142522
|
FBXO5
|
F-box protein 5
|
|
chr3_-_159306532
|
0.311
|
NM_001163678
NM_003030
NM_006884
|
SHOX2
|
short stature homeobox 2
|
|
chr3_+_52694975
|
0.310
|
NM_014366
NM_206825
NM_206826
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr7_+_43118721
|
0.310
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr17_-_69151821
|
0.309
|
NM_001144952
|
SDK2
|
sidekick homolog 2 (chicken)
|
|
chr19_+_40431618
|
0.309
|
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr11_+_75157450
|
0.307
|
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr2_+_84986273
|
0.305
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr17_+_50697319
|
0.305
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr19_-_54267690
|
0.304
|
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr3_-_107070400
|
0.304
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr2_-_60634271
|
0.303
|
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr4_+_119419354
|
0.302
|
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding)
|
|
chr7_+_156624392
|
0.302
|
NM_014671
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr1_-_142535878
|
0.302
|
|
LOC100286793
FLJ39739
|
hypothetical LOC100286793
hypothetical FLJ39739
|
|
chr19_+_40431359
|
0.301
|
NM_015925
NM_205834
NM_205835
|
LSR
|
lipolysis stimulated lipoprotein receptor
|
|
chr7_+_26298136
|
0.300
|
|
SNX10
|
sorting nexin 10
|
|
chr19_-_54267932
|
0.298
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr12_-_56111054
|
0.298
|
|
R3HDM2
|
R3H domain containing 2
|
|
chr8_+_24827173
|
0.297
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr3_+_173240983
|
0.297
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr3_+_37259684
|
0.297
|
NM_001172713
NM_002078
|
GOLGA4
|
golgin A4
|
|
chr4_+_158361185
|
0.296
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr12_-_123965539
|
0.295
|
NM_021009
|
UBC
|
ubiquitin C
|
|
chr8_-_110726201
|
0.295
|
NM_001099753
NM_001099754
NM_001099755
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr20_+_3399615
|
0.293
|
NM_139321
NM_139322
|
ATRN
|
attractin
|
|
chr2_-_88533167
|
0.293
|
NM_001135649
|
FOXI3
|
forkhead box I3
|
|
chr17_-_33043514
|
0.293
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chrX_+_150614385
|
0.293
|
NM_024082
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane)
|
|
chr10_+_105716925
|
0.293
|
|
SLK
|
STE20-like kinase
|
|
chr12_+_56230110
|
0.292
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr3_-_88190737
|
0.291
|
NM_001008390
NM_003663
|
CGGBP1
|
CGG triplet repeat binding protein 1
|
|
chr8_+_85259654
|
0.291
|
NM_001100391
|
RALYL
|
RALY RNA binding protein-like
|
|
chr12_+_120810982
|
0.290
|
NM_002813
|
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9
|
|
chr9_-_135847106
|
0.288
|
NM_001134398
NM_003371
|
VAV2
|
vav 2 guanine nucleotide exchange factor
|
|
chr2_+_68238490
|
0.287
|
NM_020143
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr3_+_53503626
|
0.287
|
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chr8_+_24827181
|
0.287
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr2_-_27195401
|
0.287
|
NM_001166240
NM_006569
|
CGREF1
|
cell growth regulator with EF-hand domain 1
|
|
chr20_-_24986501
|
0.287
|
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr14_+_101297887
|
0.286
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr3_+_40473785
|
0.286
|
NM_003973
NM_001034996
|
RPL14
|
ribosomal protein L14
|
|
chr10_-_43082372
|
0.285
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr1_+_65386359
|
0.285
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr1_-_143232275
|
0.284
|
|
LOC728855
|
hypothetical LOC728855
|
|
chr20_-_50241930
|
0.284
|
NM_018197
NM_022088
NM_199426
NM_199427
|
ZFP64
|
zinc finger protein 64 homolog (mouse)
|
|
chr6_-_28475478
|
0.283
|
NM_001163391
|
ZSCAN12
|
zinc finger and SCAN domain containing 12
|
|
chr9_-_126573349
|
0.282
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr21_+_33364316
|
0.282
|
NM_138983
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr19_-_44386731
|
0.282
|
NM_001080468
|
SYCN
|
syncollin
|
|
chr8_+_26296439
|
0.282
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr10_-_21503025
|
0.282
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr7_+_23603495
|
0.282
|
NM_138771
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr5_-_43432969
|
0.281
|
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr16_+_716936
|
0.281
|
NM_207112
NM_032304
|
HAGHL
|
hydroxyacylglutathione hydrolase-like
|
|
chr11_-_47620590
|
0.280
|
NM_014342
|
MTCH2
|
mitochondrial carrier homolog 2 (C. elegans)
|