|
chr5_-_95794416
|
2.738
|
NM_000439
|
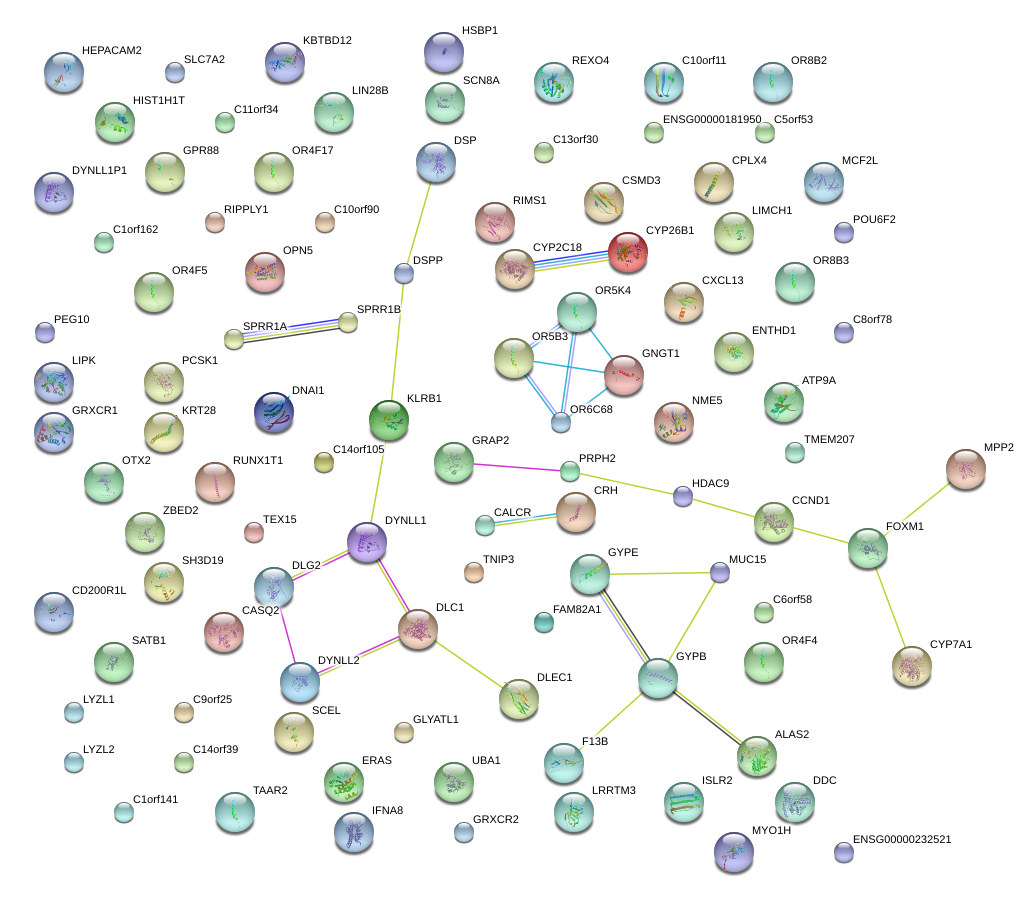
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr7_-_96471544
|
2.261
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr7_-_92693716
|
1.576
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr22_+_38652555
|
1.426
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr10_+_77212524
|
1.390
|
NM_032024
|
C10orf11
|
chromosome 10 open reading frame 11
|
|
chr14_-_60022487
|
1.382
|
NM_174978
|
C14orf39
|
chromosome 14 open reading frame 39
|
|
chr6_+_105511615
|
1.212
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr10_+_29617995
|
1.190
|
NM_032517
|
LYZL1
|
lysozyme-like 1
|
|
chr8_-_67253234
|
1.133
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr6_+_47857733
|
1.070
|
NM_181744
|
OPN5
|
opsin 5
|
|
chr6_+_7524691
|
1.052
|
|
DSP
|
desmoplakin
|
|
chr3_-_13032130
|
1.048
|
|
|
|
|
chr10_-_30958631
|
1.043
|
NM_183058
|
LYZL2
|
lysozyme-like 2
|
|
chr18_-_55136673
|
1.037
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr11_-_83071084
|
1.035
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_-_191650291
|
0.955
|
NM_207316
|
TMEM207
|
transmembrane protein 207
|
|
chr9_+_21399132
|
0.913
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chr15_+_72209973
|
0.904
|
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr6_+_127940011
|
0.901
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr3_-_114047479
|
0.891
|
NM_001008784
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr13_+_42253685
|
0.872
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr15_+_72209795
|
0.871
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr20_-_49747397
|
0.851
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr3_+_129682434
|
0.833
|
|
|
|
|
chr6_-_132980901
|
0.802
|
NM_014626
|
TAAR2
|
trace amine associated receptor 2
|
|
chr13_+_112746908
|
0.783
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr7_+_38984133
|
0.734
|
NM_001166018
NM_007252
|
POU6F2
|
POU class 6 homeobox 2
|
|
chrX_-_106033202
|
0.711
|
NM_001171706
NM_138382
|
RIPPLY1
|
ripply1 homolog (zebrafish)
|
|
chr17_-_36209672
|
0.700
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr7_+_93373755
|
0.674
|
NM_021955
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1
|
|
chr7_+_18296569
|
0.663
|
|
HDAC9
|
histone deacetylase 9
|
|
chr8_-_125252884
|
0.657
|
|
C8orf78
|
chromosome 8 open reading frame 78
|
|
chr11_-_26550243
|
0.628
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr5_-_137502975
|
0.628
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr10_-_128199986
|
0.626
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr1_+_111818111
|
0.625
|
NM_174896
|
C1orf162
|
chromosome 1 open reading frame 162
|
|
chr1_-_195302973
|
0.616
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr6_-_26216299
|
0.614
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr1_-_116112684
|
0.607
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr4_-_145281237
|
0.606
|
NM_002099
|
GYPA
|
glycophorin A (MNS blood group)
|
|
chr15_-_100280784
|
0.603
|
NM_001004195
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr3_+_99555387
|
0.600
|
NM_001005517
|
OR5K4
|
olfactory receptor, family 5, subfamily K, member 4
|
|
chr8_-_114518194
|
0.593
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr14_-_56342097
|
0.586
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr9_-_34448477
|
0.581
|
NM_001184940
NM_001184941
NM_001184942
NM_001184943
NM_001184944
NM_001184945
NM_147202
|
C9orf25
|
chromosome 9 open reading frame 25
|
|
chr1_-_116112853
|
0.574
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr6_+_72653126
|
0.570
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr10_+_96433240
|
0.565
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr4_+_78651929
|
0.555
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr22_-_38619739
|
0.546
|
NM_152512
|
ENTHD1
|
ENTH domain containing 1
|
|
chr14_-_57030227
|
0.544
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr1_-_116112921
|
0.543
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr9_+_34448810
|
0.540
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr8_+_17440664
|
0.539
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr4_-_145159889
|
0.527
|
NM_002100
|
GYPB
|
glycophorin B (MNS blood group)
|
|
chr10_+_68355797
|
0.513
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr3_-_18455207
|
0.510
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr1_+_58899
|
0.504
|
NM_001005484
|
OR4F4
OR4F5
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 5
|
|
chr4_+_42590038
|
0.499
|
NM_001080476
|
GRXCR1
|
glutaredoxin, cysteine rich 1
|
|
chr3_+_129124591
|
0.498
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr6_+_72983183
|
0.492
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr11_+_69176391
|
0.491
|
|
CCND1
|
cyclin D1
|
|
chr19_+_61624
|
0.486
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr4_-_122304925
|
0.485
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr22_+_21116283
|
0.484
|
|
|
|
|
chr8_-_13018123
|
0.477
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr6_+_72982865
|
0.468
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_-_67366807
|
0.459
|
NM_001013674
|
C1orf141
|
chromosome 1 open reading frame 141
|
|
chr7_+_94135384
|
0.457
|
|
PEG10
|
paternally expressed 10
|
|
chr12_+_108310906
|
0.454
|
NM_001101421
|
MYO1H
|
myosin IH
|
|
chr10_+_90474280
|
0.437
|
NM_001080518
|
LIPK
|
lipase, family member K
|
|
chr7_-_93027057
|
0.436
|
NM_001164738
|
CALCR
|
calcitonin receptor
|
|
chrX_+_48572226
|
0.435
|
NM_181532
|
ERAS
|
ES cell expressed Ras
|
|
chr11_-_123758448
|
0.435
|
NM_001005468
|
OR8B2
|
olfactory receptor, family 8, subfamily B, member 2
|
|
chr5_+_139485703
|
0.424
|
NM_001007189
|
C5orf53
|
chromosome 5 open reading frame 53
|
|
chr1_+_151270301
|
0.419
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr8_-_93181091
|
0.412
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_30826074
|
0.406
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr11_-_57927457
|
0.401
|
NM_001005469
|
OR5B3
|
olfactory receptor, family 5, subfamily B, member 3
|
|
chr11_+_58467297
|
0.393
|
NM_080661
|
GLYATL1
|
glycine-N-acyltransferase-like 1
|
|
chr1_-_120189301
|
0.391
|
NM_001047980
|
NBPF7
|
neuroblastoma breakpoint family, member 7
|
|
chr13_+_77007809
|
0.388
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr11_-_111636792
|
0.388
|
NM_001145024
|
C11orf34
|
chromosome 11 open reading frame 34
|
|
chrX_-_55074125
|
0.385
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr2_+_38009405
|
0.382
|
NM_001170792
|
FAM82A1
|
family with sequence similarity 82, member A1
|
|
chr4_-_152368492
|
0.379
|
|
SH3D19
|
SH3 domain containing 19
|
|
chr7_-_50596238
|
0.376
|
NM_000790
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr5_-_145232723
|
0.376
|
NM_001080516
|
GRXCR2
|
glutaredoxin, cysteine rich 2
|
|
chr12_+_54172413
|
0.372
|
NM_001005519
|
OR6C68
|
olfactory receptor, family 6, subfamily C, member 68
|
|
chr4_+_41309668
|
0.366
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_100776280
|
0.365
|
NM_022049
|
GPR88
|
G protein-coupled receptor 88
|
|
chr3_-_112796855
|
0.360
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr12_+_50271283
|
0.359
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chrX_+_46938215
|
0.356
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr4_+_41309482
|
0.341
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr8_-_59575249
|
0.338
|
NM_000780
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1
|
|
chr2_-_72224696
|
0.332
|
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chrX_+_28515479
|
0.331
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr9_+_19280748
|
0.331
|
NM_017925
|
DENND4C
|
DENN/MADD domain containing 4C
|
|
chr9_-_124981125
|
0.328
|
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr2_+_27652892
|
0.325
|
NM_032266
|
C2orf16
|
chromosome 2 open reading frame 16
|
|
chr1_+_178466064
|
0.320
|
NM_033343
|
LHX4
|
LIM homeobox 4
|
|
chr12_+_20854904
|
0.318
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr5_-_75954807
|
0.317
|
NM_004101
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2
|
|
chr10_-_17211795
|
0.311
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr11_-_11331411
|
0.307
|
|
CSNK2A1P
|
casein kinase 2, alpha 1 polypeptide pseudogene
|
|
chr3_+_143858428
|
0.306
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr7_+_133862883
|
0.306
|
NM_020299
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase)
|
|
chrX_+_46938136
|
0.305
|
NM_003334
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr1_-_229071957
|
0.305
|
NM_001136494
|
C1orf198
|
chromosome 1 open reading frame 198
|
|
chr4_+_41309690
|
0.303
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr7_+_72034735
|
0.301
|
|
POM121
|
POM121 membrane glycoprotein
|
|
chr7_-_83116414
|
0.298
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr3_-_152641248
|
0.295
|
NM_001178145
NM_001178146
|
IGSF10
|
immunoglobulin superfamily, member 10
|
|
chr2_-_138762909
|
0.295
|
|
|
|
|
chr1_-_161380562
|
0.294
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr3_+_187813553
|
0.293
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr13_-_45577144
|
0.291
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr2_+_158666627
|
0.290
|
NM_173355
|
UPP2
|
uridine phosphorylase 2
|
|
chr9_+_80134302
|
0.289
|
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chrX_+_46938171
|
0.283
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr1_-_158815895
|
0.282
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr18_+_59295123
|
0.280
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr9_-_21177529
|
0.279
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr1_+_61642287
|
0.276
|
|
NFIA
|
nuclear factor I/A
|
|
chr7_-_38279757
|
0.274
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr3_+_131761867
|
0.272
|
NM_001102608
|
COL6A6
|
collagen, type VI, alpha 6
|
|
chr8_-_91164100
|
0.270
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr13_-_25687062
|
0.266
|
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr8_-_124818754
|
0.264
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr2_-_21081694
|
0.264
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr16_-_3291378
|
0.260
|
NM_033208
|
TIGD7
|
tigger transposable element derived 7
|
|
chr1_+_158603480
|
0.260
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr1_-_76170563
|
0.259
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr2_-_180319013
|
0.258
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chr21_-_26269298
|
0.258
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr12_+_52126131
|
0.256
|
|
PRR13
|
proline rich 13
|
|
chr1_+_160599396
|
0.248
|
NM_001126060
|
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein
|
|
chr3_+_46717826
|
0.245
|
NM_147196
|
TMIE
|
transmembrane inner ear
|
|
chr1_+_67545634
|
0.244
|
NM_001559
|
IL12RB2
|
interleukin 12 receptor, beta 2
|
|
chrX_+_41433169
|
0.241
|
NM_001097579
NM_005300
|
GPR34
|
G protein-coupled receptor 34
|
|
chr4_+_22303645
|
0.240
|
NM_001128432
NM_020973
|
GBA3
|
glucosidase, beta, acid 3 (cytosolic)
|
|
chr6_+_132933153
|
0.239
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chrX_+_46938157
|
0.237
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr11_-_58736897
|
0.236
|
NM_001039396
|
MPEG1
|
macrophage expressed 1
|
|
chr3_+_35658852
|
0.235
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr14_-_20098929
|
0.233
|
NM_001001673
NM_001110356
NM_001110357
NM_001110358
NM_001110359
NM_001110360
NM_001110361
|
RNASE9
|
ribonuclease, RNase A family, 9 (non-active)
|
|
chr5_+_92944680
|
0.230
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr19_+_24061717
|
0.227
|
NM_203282
|
ZNF254
|
zinc finger protein 254
|
|
chr11_+_22646229
|
0.221
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr12_+_21175394
|
0.221
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr5_-_145503190
|
0.218
|
|
LARS
|
leucyl-tRNA synthetase
|
|
chr7_+_120490054
|
0.218
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr10_+_83627422
|
0.217
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr5_+_125786717
|
0.215
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr3_+_99350859
|
0.214
|
NM_001005514
|
OR5H14
|
olfactory receptor, family 5, subfamily H, member 14
|
|
chr1_-_116184846
|
0.214
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr3_+_162697289
|
0.213
|
NM_001080440
|
OTOL1
|
otolin 1 homolog (zebrafish)
|
|
chr18_-_58946858
|
0.213
|
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr17_+_46176105
|
0.210
|
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr19_-_22171592
|
0.209
|
NM_001001411
|
ZNF676
|
zinc finger protein 676
|
|
chrX_+_46938178
|
0.209
|
|
UBA1
|
ubiquitin-like modifier activating enzyme 1
|
|
chr17_+_17548888
|
0.208
|
|
|
|
|
chr15_-_48626193
|
0.205
|
NM_203494
|
USP50
|
ubiquitin specific peptidase 50
|
|
chr4_-_24641404
|
0.203
|
NM_018176
|
LGI2
|
leucine-rich repeat LGI family, member 2
|
|
chr4_+_35962038
|
0.201
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr14_-_105623358
|
0.198
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr18_-_28606839
|
0.197
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr11_-_56166490
|
0.196
|
NM_001002925
|
OR5AP2
|
olfactory receptor, family 5, subfamily AP, member 2
|
|
chr1_+_234748187
|
0.195
|
NM_201545
|
LGALS8
|
lectin, galactoside-binding, soluble, 8
|
|
chr6_+_30238961
|
0.195
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr4_+_74497998
|
0.194
|
|
ALB
|
albumin
|
|
chr1_-_150044365
|
0.194
|
NM_001004432
|
LINGO4
|
leucine rich repeat and Ig domain containing 4
|
|
chr1_-_214963279
|
0.193
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr3_+_192413015
|
0.192
|
NM_198184
|
OSTN
|
osteocrin
|
|
chr2_+_161701711
|
0.192
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr19_-_51236113
|
0.191
|
NM_001002923
|
IGFL4
|
IGF-like family member 4
|
|
chr20_+_71230
|
0.190
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr7_+_142539291
|
0.190
|
NM_002652
|
PIP
|
prolactin-induced protein
|
|
chr2_-_121964373
|
0.189
|
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr16_+_4842077
|
0.189
|
NM_016936
|
UBN1
|
ubinuclein 1
|
|
chr5_+_161207262
|
0.186
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr12_-_4358968
|
0.186
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chrX_+_150635162
|
0.186
|
NM_033085
|
FATE1
|
fetal and adult testis expressed 1
|
|
chr8_+_80685927
|
0.186
|
|
STMN2
|
stathmin-like 2
|
|
chr11_+_122214415
|
0.185
|
NM_019604
|
CRTAM
|
cytotoxic and regulatory T cell molecule
|
|
chr8_+_80685931
|
0.183
|
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr9_+_35031101
|
0.182
|
NM_001040411
NM_001040412
|
C9orf131
|
chromosome 9 open reading frame 131
|
|
chr7_-_27125738
|
0.182
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr1_+_66230977
|
0.181
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr12_-_11106159
|
0.180
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr2_+_88966170
|
0.177
|
|
|
|
|
chr1_-_34404029
|
0.176
|
NM_052896
|
CSMD2
|
CUB and Sushi multiple domains 2
|
|
chr11_-_59390518
|
0.176
|
NM_001062
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family)
|
|
chr5_+_176449156
|
0.175
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chrX_+_135106578
|
0.172
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr4_+_74498042
|
0.169
|
|
ALB
|
albumin
|
|
chr11_+_5466490
|
0.169
|
NM_001005163
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1
|
|
chr11_-_123772456
|
0.168
|
NM_001005467
|
OR8B3
|
olfactory receptor, family 8, subfamily B, member 3
|
|
chr17_-_44400952
|
0.167
|
NM_004123
|
GIP
|
gastric inhibitory polypeptide
|