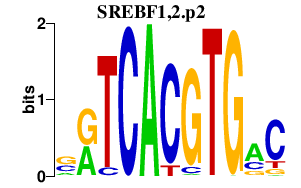
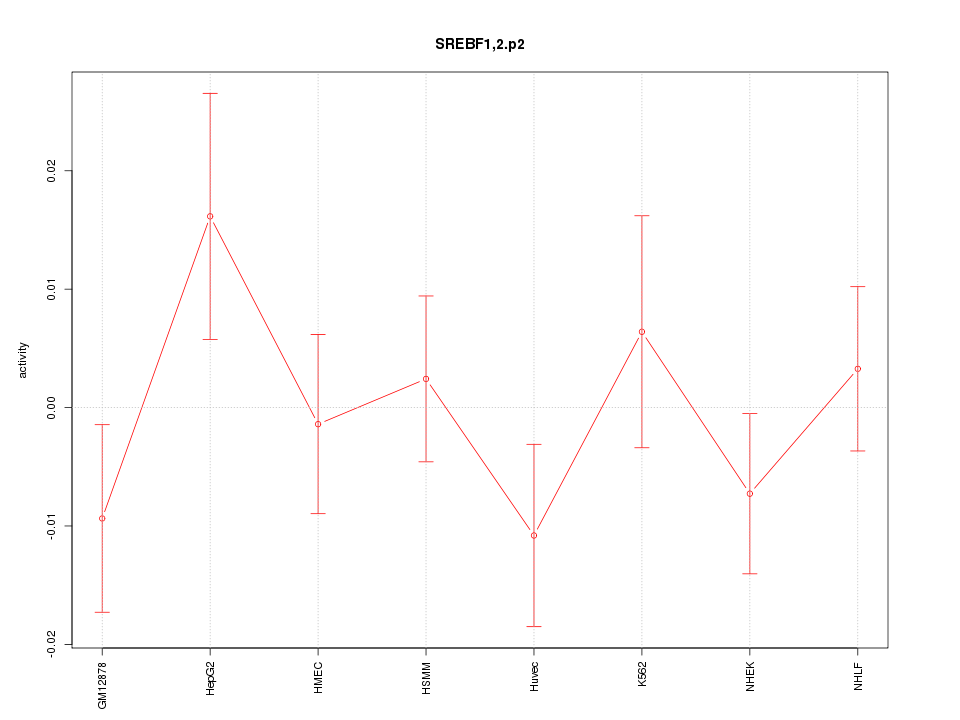
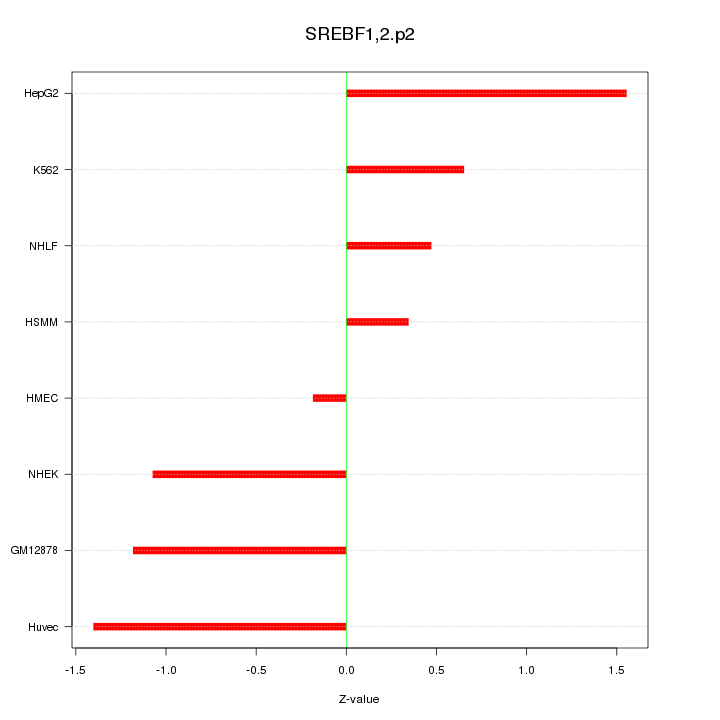
|
chr13_+_48582389
|
1.800
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr2_+_234333653
|
1.578
|
NM_000463
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chr2_+_227898163
|
1.445
|
|
MFF
|
mitochondrial fission factor
|
|
chr20_+_37024383
|
1.372
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr14_-_34253473
|
1.312
|
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr2_+_177785667
|
1.305
|
NM_194247
|
HNRNPA3P1
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 pseudogene 1
heterogeneous nuclear ribonucleoprotein A3
|
|
chr2_-_176575171
|
1.160
|
NM_030650
|
KIAA1715
|
KIAA1715
|
|
chr6_-_84994032
|
1.155
|
NM_014895
|
KIAA1009
|
KIAA1009
|
|
chr10_-_45410197
|
1.092
|
NM_001002265
NM_145021
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr17_-_44022595
|
1.070
|
|
HOXB3
|
homeobox B3
|
|
chr10_+_45542653
|
1.059
|
NM_001169106
NM_001169107
NM_015262
|
FAM21C
FAM21A
|
family with sequence similarity 21, member C
family with sequence similarity 21, member A
|
|
chr11_-_85457469
|
1.027
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr6_-_7258351
|
1.001
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr6_+_35852369
|
0.993
|
NM_207409
|
C6orf126
|
chromosome 6 open reading frame 126
|
|
chr11_+_85017269
|
0.990
|
NM_001193537
NM_001193538
NM_018480
|
TMEM126B
|
transmembrane protein 126B
|
|
chr14_-_67353046
|
0.978
|
NM_015346
|
ZFYVE26
|
zinc finger, FYVE domain containing 26
|
|
chr20_-_43953036
|
0.975
|
NM_080749
|
NEURL2
|
neuralized homolog 2 (Drosophila)
|
|
chr17_-_41018291
|
0.968
|
|
|
|
|
chr16_+_55580910
|
0.964
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr20_+_43953428
|
0.909
|
|
CTSA
|
cathepsin A
|
|
chr2_-_131567458
|
0.884
|
NM_001009993
|
FAM168B
|
family with sequence similarity 168, member B
|
|
chr20_+_43953372
|
0.882
|
|
CTSA
|
cathepsin A
|
|
chr2_+_187266899
|
0.879
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr3_-_20202622
|
0.868
|
NM_001012409
NM_001012410
NM_001012411
NM_001012412
NM_001012413
NM_138484
|
SGOL1
|
shugoshin-like 1 (S. pombe)
|
|
chr6_-_169392771
|
0.867
|
|
|
|
|
chr10_+_69314714
|
0.860
|
|
SIRT1
|
sirtuin 1
|
|
chr11_+_125586828
|
0.858
|
NM_024556
|
FAM118B
|
family with sequence similarity 118, member B
|
|
chr2_+_176724583
|
0.851
|
|
|
|
|
chr2_-_27399405
|
0.848
|
NM_002437
|
MPV17
|
MpV17 mitochondrial inner membrane protein
|
|
chr3_-_11736946
|
0.845
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr6_+_160310118
|
0.843
|
NM_000876
|
IGF2R
|
insulin-like growth factor 2 receptor
|
|
chr6_+_127629677
|
0.828
|
NM_030963
|
RNF146
|
ring finger protein 146
|
|
chr7_-_44496703
|
0.824
|
NM_015332
|
NUDCD3
|
NudC domain containing 3
|
|
chr4_-_23500706
|
0.821
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr2_+_241174803
|
0.816
|
NM_023083
NM_023085
|
CAPN10
|
calpain 10
|
|
chr20_+_33506322
|
0.808
|
NM_007186
|
CEP250
|
centrosomal protein 250kDa
|
|
chr1_+_224337937
|
0.807
|
|
|
|
|
chr7_-_134845363
|
0.802
|
NM_001008225
NM_001190847
NM_001190848
NM_001190849
NM_001190850
NM_013316
|
CNOT4
|
CCR4-NOT transcription complex, subunit 4
|
|
chr2_+_176724358
|
0.779
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr2_+_85676347
|
0.777
|
NM_016494
|
RNF181
|
ring finger protein 181
|
|
chr16_+_68542126
|
0.774
|
NM_001136214
|
CLEC18A
|
C-type lectin domain family 18, member A
|
|
chr6_+_139391511
|
0.773
|
NM_021243
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr20_+_43953331
|
0.768
|
|
CTSA
|
cathepsin A
|
|
chr13_+_112999590
|
0.760
|
|
LAMP1
|
lysosomal-associated membrane protein 1
|
|
chr22_+_34107087
|
0.749
|
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr1_-_152797820
|
0.748
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr5_+_41940216
|
0.730
|
NM_175921
|
C5orf51
|
chromosome 5 open reading frame 51
|
|
chr16_+_68542310
|
0.724
|
NM_182619
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr13_-_78131259
|
0.704
|
NM_024546
|
RNF219
|
ring finger protein 219
|
|
chr1_+_76312962
|
0.700
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr16_-_1465007
|
0.686
|
|
CLCN7
|
chloride channel 7
|
|
chr16_-_1465041
|
0.685
|
NM_001114331
NM_001287
|
CLCN7
|
chloride channel 7
|
|
chr6_+_7671267
|
0.684
|
|
BMP6
|
bone morphogenetic protein 6
|
|
chr1_+_159550789
|
0.683
|
NM_001035511
NM_001035512
NM_001035513
NM_003001
|
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa
|
|
chr20_+_33823318
|
0.676
|
NM_016436
|
PHF20
|
PHD finger protein 20
|
|
chr1_-_152797707
|
0.673
|
NM_017582
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr19_+_10626106
|
0.668
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr11_-_85457736
|
0.662
|
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr11_-_85457753
|
0.658
|
NM_001008660
NM_007166
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr6_+_151815298
|
0.655
|
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr16_+_5023672
|
0.647
|
|
LOC100507589
|
hypothetical LOC100507589
|
|
chr4_+_17225344
|
0.644
|
NM_025205
|
MED28
|
mediator complex subunit 28
|
|
chr1_-_152797760
|
0.639
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr6_+_139391570
|
0.639
|
|
C6orf115
|
chromosome 6 open reading frame 115
|
|
chr5_+_133735004
|
0.635
|
|
UBE2B
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog)
|
|
chr20_-_32877029
|
0.635
|
NM_014071
|
NCOA6
|
nuclear receptor coactivator 6
|
|
chr16_-_15644465
|
0.627
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr1_-_152797767
|
0.621
|
|
UBE2Q1
|
ubiquitin-conjugating enzyme E2Q family member 1
|
|
chr20_+_46971618
|
0.615
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr16_-_5023562
|
0.614
|
|
NAGPA
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr16_-_1464927
|
0.595
|
|
CLCN7
|
chloride channel 7
|
|
chr6_-_163754903
|
0.594
|
|
LOC100526820
|
hypothetical LOC100526820
|
|
chr7_+_27190688
|
0.594
|
|
|
|
|
chr7_-_135083906
|
0.592
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr16_+_28893525
|
0.592
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr20_+_18066473
|
0.590
|
NM_001164811
|
PET117
|
cytochrome c oxidase assembly factor-like
|
|
chr6_-_21631992
|
0.587
|
|
|
|
|
chr15_-_71712605
|
0.587
|
|
NPTN
|
neuroplastin
|
|
chr14_-_23981493
|
0.585
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr3_+_87359088
|
0.573
|
NM_014043
|
CHMP2B
|
chromatin modifying protein 2B
|
|
chr6_-_26767892
|
0.570
|
NM_024639
|
ZNF322A
|
zinc finger protein 322A
|
|
chr8_+_98725505
|
0.570
|
NM_178812
|
MTDH
|
metadherin
|
|
chr17_-_44043361
|
0.569
|
NM_004502
|
HOXB7
|
homeobox B7
|
|
chr8_-_54918099
|
0.568
|
NM_213620
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H
|
|
chr17_-_43977273
|
0.564
|
NM_002145
|
HOXB2
|
homeobox B2
|
|
chr13_-_32757835
|
0.564
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_-_27210330
|
0.563
|
NM_018405
|
C17orf79
|
chromosome 17 open reading frame 79
|
|
chr16_+_80370426
|
0.558
|
NM_002661
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr1_+_10015736
|
0.555
|
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chr16_-_1465021
|
0.554
|
|
CLCN7
|
chloride channel 7
|
|
chr1_-_201162991
|
0.551
|
NM_021633
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr22_+_38903825
|
0.549
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr1_+_234372454
|
0.544
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr2_+_46779827
|
0.541
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr14_-_23752507
|
0.531
|
|
CHMP4A
|
chromatin modifying protein 4A
|
|
chr16_+_22124870
|
0.529
|
NM_013302
|
EEF2K
|
eukaryotic elongation factor-2 kinase
|
|
chr14_+_30161267
|
0.528
|
NM_016106
NM_182835
|
SCFD1
|
sec1 family domain containing 1
|
|
chr1_+_109221271
|
0.528
|
|
GPSM2
|
G-protein signaling modulator 2
|
|
chr3_+_150330060
|
0.527
|
NM_032383
|
HPS3
|
Hermansky-Pudlak syndrome 3
|
|
chr16_+_83618804
|
0.519
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr8_-_95343716
|
0.516
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr3_-_126257425
|
0.515
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr17_-_40263124
|
0.514
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr10_+_69314347
|
0.511
|
NM_012238
|
SIRT1
|
sirtuin 1
|
|
chr1_+_109963982
|
0.510
|
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr2_-_200424140
|
0.509
|
|
LOC348751
|
hypothetical LOC348751
|
|
chr14_-_23752476
|
0.506
|
|
CHMP4A
|
chromatin modifying protein 4A
|
|
chr20_-_61317891
|
0.505
|
NM_017798
|
YTHDF1
|
YTH domain family, member 1
|
|
chr14_-_23752498
|
0.505
|
|
CHMP4A
|
chromatin modifying protein 4A
|
|
chr19_-_13088212
|
0.504
|
NM_001142554
NM_017722
NM_001136035
|
TRMT1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr4_-_84152985
|
0.499
|
NM_001115007
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr17_-_44042924
|
0.498
|
|
HOXB7
|
homeobox B7
|
|
chr1_+_149750485
|
0.494
|
NM_020770
|
CGN
|
cingulin
|
|
chr6_-_31973418
|
0.493
|
|
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2
|
|
chr1_+_181871830
|
0.491
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr9_-_130749604
|
0.490
|
NM_014908
|
DOLK
|
dolichol kinase
|
|
chr19_-_15436333
|
0.489
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr17_+_52410466
|
0.489
|
NM_021626
|
SCPEP1
|
serine carboxypeptidase 1
|
|
chr2_-_148494732
|
0.488
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr2_-_196744546
|
0.487
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr17_-_34263487
|
0.487
|
NM_000978
|
SNORA21
RPL23
|
small nucleolar RNA, H/ACA box 21
ribosomal protein L23
|
|
chr1_-_201162938
|
0.479
|
|
KLHL12
|
kelch-like 12 (Drosophila)
|
|
chr2_-_105312536
|
0.479
|
NM_001142621
NM_004257
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1
|
|
chr10_-_120504659
|
0.475
|
NM_153810
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr6_-_43305156
|
0.475
|
NM_006443
NM_199184
|
C6orf108
|
chromosome 6 open reading frame 108
|
|
chr20_-_4743768
|
0.474
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr1_-_77920914
|
0.472
|
NM_015534
|
ZZZ3
|
zinc finger, ZZ-type containing 3
|
|
chr20_+_43952997
|
0.472
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr2_+_46779553
|
0.468
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr17_-_73636386
|
0.467
|
NM_001127198
|
TMC6
|
transmembrane channel-like 6
|
|
chr14_-_38971370
|
0.465
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr10_-_120504445
|
0.465
|
|
C10orf46
|
chromosome 10 open reading frame 46
|
|
chr12_-_47396724
|
0.463
|
NM_001240
|
CCNT1
|
cyclin T1
|
|
chr2_+_223625105
|
0.463
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr20_+_18496068
|
0.461
|
|
LOC388789
|
hypothetical LOC388789
|
|
chr8_-_95343696
|
0.459
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr11_-_125586733
|
0.458
|
NM_001144827
NM_032795
|
RPUSD4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr22_+_34107051
|
0.456
|
NM_002133
|
HMOX1
|
heme oxygenase (decycling) 1
|
|
chr5_-_55564942
|
0.452
|
NM_024669
|
ANKRD55
|
ankyrin repeat domain 55
|
|
chr17_-_44043278
|
0.450
|
|
HOXB7
|
homeobox B7
|
|
chr8_-_81246215
|
0.447
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr16_+_80036275
|
0.447
|
NM_198390
|
CMIP
|
c-Maf-inducing protein
|
|
chr14_+_37746937
|
0.445
|
NM_001049
|
SSTR1
|
somatostatin receptor 1
|
|
chr6_-_31973434
|
0.444
|
NM_006709
NM_025256
|
EHMT2
|
euchromatic histone-lysine N-methyltransferase 2
|
|
chr17_-_33043514
|
0.443
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr10_+_101532544
|
0.441
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr8_+_98725604
|
0.440
|
|
MTDH
|
metadherin
|
|
chr17_-_7078304
|
0.438
|
|
DVL2
|
dishevelled, dsh homolog 2 (Drosophila)
|
|
chr12_+_54396118
|
0.438
|
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr7_+_97574054
|
0.437
|
NM_014916
|
LMTK2
|
lemur tyrosine kinase 2
|
|
chr2_-_210743622
|
0.436
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr17_+_63252704
|
0.435
|
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr6_+_87921980
|
0.434
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr5_+_122138636
|
0.432
|
NM_003100
|
SNX2
|
sorting nexin 2
|
|
chr5_-_172130766
|
0.430
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr14_+_22860511
|
0.430
|
|
PABPN1
|
poly(A) binding protein, nuclear 1
|
|
chr6_-_44333002
|
0.429
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr4_-_152901542
|
0.428
|
NM_004564
|
PET112L
|
PET112-like (yeast)
|
|
chr11_-_1741680
|
0.428
|
|
CTSD
|
cathepsin D
|
|
chr17_-_73636362
|
0.427
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr1_+_10015577
|
0.424
|
NM_001105562
NM_006048
|
UBE4B
|
ubiquitination factor E4B (UFD2 homolog, yeast)
|
|
chr14_-_34661190
|
0.423
|
NM_017917
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr17_+_63252087
|
0.423
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr16_+_15644624
|
0.422
|
NM_001143979
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr17_-_71362971
|
0.419
|
NM_012478
|
WBP2
|
WW domain binding protein 2
|
|
chr1_+_109963950
|
0.419
|
NM_139156
|
AMPD2
|
adenosine monophosphate deaminase 2
|
|
chr6_+_151815114
|
0.419
|
NM_024573
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr16_+_4837912
|
0.419
|
NM_001079514
|
UBN1
|
ubinuclein 1
|
|
chr6_-_33822659
|
0.418
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr16_+_80370400
|
0.417
|
|
PLCG2
|
phospholipase C, gamma 2 (phosphatidylinositol-specific)
|
|
chr3_+_4996096
|
0.417
|
NM_003670
|
BHLHE40
|
basic helix-loop-helix family, member e40
|
|
chr18_-_20231709
|
0.415
|
|
OSBPL1A
|
oxysterol binding protein-like 1A
|
|
chr1_+_36462583
|
0.412
|
NM_005119
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr16_-_57276108
|
0.412
|
NM_018231
|
SLC38A7
|
solute carrier family 38, member 7
|
|
chr10_+_101532450
|
0.410
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr9_-_124707314
|
0.409
|
NM_001100588
NM_018835
|
RC3H2
|
ring finger and CCCH-type domains 2
|
|
chr20_-_34102250
|
0.407
|
|
LOC647979
|
hypothetical LOC647979
|
|
chr11_-_8849485
|
0.406
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr6_-_152664906
|
0.406
|
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_-_139128777
|
0.405
|
|
DPP7
|
dipeptidyl-peptidase 7
|
|
chr14_-_93512818
|
0.404
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr14_-_34661156
|
0.403
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr2_-_175741096
|
0.402
|
NM_001880
|
ATF2
|
activating transcription factor 2
|
|
chr12_-_119826073
|
0.400
|
|
SPPL3
|
signal peptide peptidase 3
|
|
chr2_+_68238490
|
0.398
|
NM_020143
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr2_-_215605054
|
0.396
|
NM_015657
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr16_-_87450734
|
0.395
|
|
GALNS
|
galactosamine (N-acetyl)-6-sulfate sulfatase
|
|
chr11_+_66915997
|
0.393
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr9_-_135379888
|
0.392
|
NM_001080483
|
TMEM8C
|
transmembrane protein 8C
|
|
chr12_+_70519753
|
0.391
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr16_+_66855827
|
0.389
|
NM_001076785
NM_003983
|
SLC7A6
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 6
|
|
chr10_-_120504267
|
0.389
|
|
|
|
|
chr1_+_36462625
|
0.387
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr1_-_154532019
|
0.387
|
NM_144580
|
C1orf85
|
chromosome 1 open reading frame 85
|
|
chr5_+_122138661
|
0.387
|
|
SNX2
|
sorting nexin 2
|
|
chr17_+_46599081
|
0.386
|
NM_001018138
|
NME2
|
non-metastatic cells 2, protein (NM23B) expressed in
|
|
chr16_+_11669736
|
0.386
|
NM_003498
|
SNN
|
stannin
|
|
chr15_-_32398220
|
0.383
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr12_+_102982365
|
0.382
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr11_-_6597220
|
0.380
|
|
TPP1
|
tripeptidyl peptidase I
|