|
chr6_+_36752244
|
3.569
|
NM_078467
|
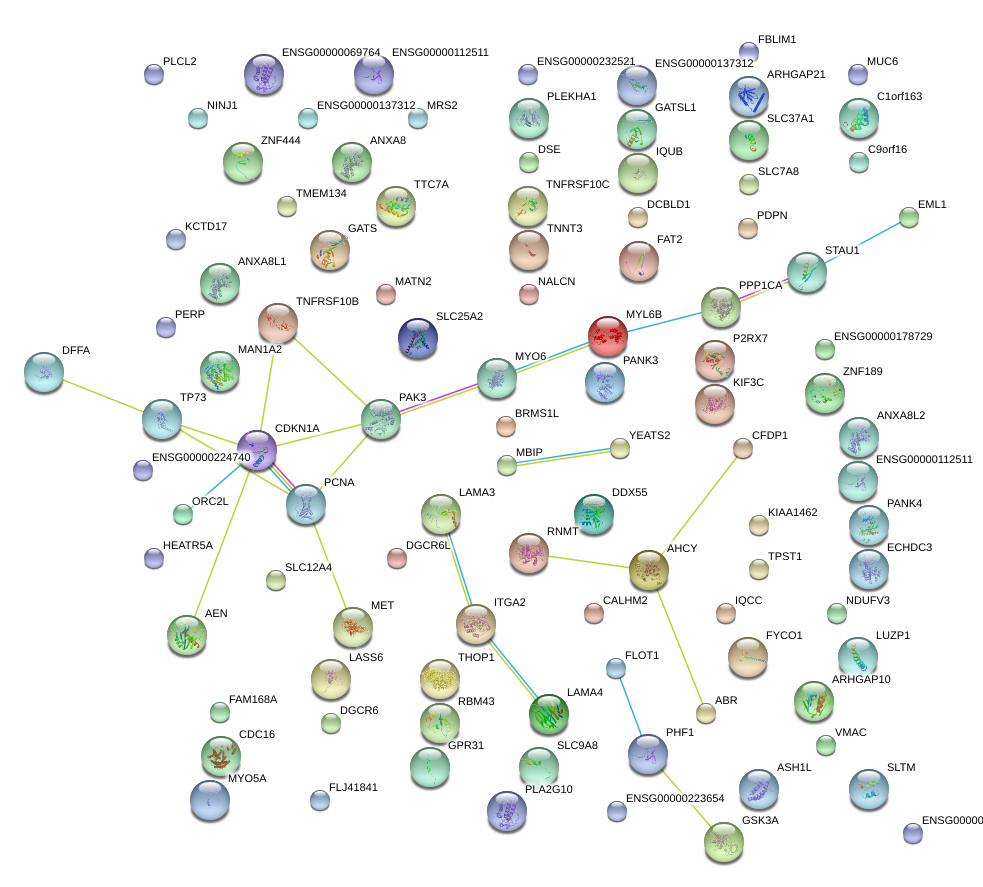
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr1_+_3597092
|
1.655
|
NM_001126240
NM_001126241
NM_001126242
|
TP73
|
tumor protein p73
|
|
chr6_-_138470160
|
1.356
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr16_-_66558197
|
1.266
|
NM_001145964
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr6_+_24511123
|
1.089
|
NM_020662
|
MRS2
|
MRS2 magnesium homeostasis factor homolog (S. cerevisiae)
|
|
chr15_-_50608296
|
1.050
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr1_-_2447848
|
0.968
|
NM_018216
|
PANK4
|
pantothenate kinase 4
|
|
chr6_+_116798791
|
0.961
|
NM_013352
|
DSE
|
dermatan sulfate epimerase
|
|
chr22_-_18687511
|
0.948
|
NM_033257
|
DGCR6L
|
DiGeorge syndrome critical region gene 6-like
|
|
chr11_-_72986738
|
0.890
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr10_-_30388439
|
0.890
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr7_-_74705276
|
0.885
|
NM_001145064
|
GATSL2
|
GATS protein-like 2
|
|
chr6_-_167491305
|
0.787
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr1_+_153799637
|
0.763
|
|
LOC645676
|
hypothetical LOC645676
|
|
chr1_+_32443822
|
0.753
|
NM_001160042
NM_018134
|
IQCC
|
IQ motif containing C
|
|
chr20_-_32354736
|
0.746
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr14_-_30959487
|
0.717
|
|
HEATR5A
|
HEAT repeat containing 5A
|
|
chr2_-_26058941
|
0.707
|
NM_002254
|
KIF3C
|
kinesin family member 3C
|
|
chr16_-_14695930
|
0.637
|
NM_003561
|
PLA2G10
|
phospholipase A2, group X
|
|
chr4_+_148872902
|
0.636
|
NM_024605
|
ARHGAP10
|
Rho GTPase activating protein 10
|
|
chr22_+_35777561
|
0.617
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chr5_+_52320660
|
0.617
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr7_+_116099648
|
0.605
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr9_+_129962292
|
0.593
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chr7_+_65307816
|
0.589
|
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr19_+_5855842
|
0.584
|
NM_001017921
|
VMAC
|
vimentin-type intermediate filament associated coiled-coil protein
|
|
chr19_+_2736464
|
0.568
|
NM_003249
|
THOP1
|
thimet oligopeptidase 1
|
|
chr2_+_169021001
|
0.548
|
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr12_+_122652572
|
0.548
|
NM_020936
|
DDX55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55
|
|
chr6_+_117910472
|
0.544
|
NM_173674
|
DCBLD1
|
discoidin, CUB and LCCL domain containing 1
|
|
chr8_+_23016271
|
0.542
|
NM_003841
|
TNFRSF10C
|
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain
|
|
chr2_+_47021866
|
0.540
|
|
TTC7A
|
tetratricopeptide repeat domain 7A
|
|
chr2_-_26058831
|
0.534
|
|
KIF3C
|
kinesin family member 3C
|
|
chr2_+_169021039
|
0.513
|
NM_203463
|
LASS6
|
LAG1 homolog, ceramide synthase 6
|
|
chr3_+_184898234
|
0.510
|
NM_018023
|
YEATS2
|
YEATS domain containing 2
|
|
chr21_+_42807052
|
0.490
|
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr2_+_47021816
|
0.474
|
NM_020458
|
TTC7A
|
tetratricopeptide repeat domain 7A
|
|
chr15_+_86965526
|
0.473
|
NM_022767
|
AEN
|
apoptosis enhancing nuclease
|
|
chr9_-_94936319
|
0.465
|
NM_004148
|
NINJ1
|
ninjurin 1
|
|
chr19_-_47438583
|
0.465
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr2_-_201536425
|
0.463
|
NM_006190
|
ORC2
|
origin recognition complex, subunit 2
|
|
chr19_+_61344501
|
0.444
|
|
ZNF444
|
zinc finger protein 444
|
|
chr7_+_72206961
|
0.440
|
|
GTF2IP1
|
general transcription factor IIi, pseudogene 1
|
|
chr1_+_15957832
|
0.432
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr8_+_98950453
|
0.432
|
NM_002380
NM_030583
|
MATN2
|
matrilin 2
|
|
chr1_-_52936573
|
0.431
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr20_+_47862782
|
0.428
|
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr14_+_99273751
|
0.426
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr19_-_47438384
|
0.424
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr10_-_105202139
|
0.423
|
NM_015916
|
CALHM2
|
calcium homeostasis modulator 2
|
|
chr14_+_35365266
|
0.422
|
NM_032352
|
BRMS1L
|
breast cancer metastasis-suppressor 1-like
|
|
chr12_+_54832470
|
0.421
|
NM_002475
|
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle
|
|
chr1_+_13782838
|
0.417
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr19_-_47438563
|
0.415
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr20_+_47862656
|
0.414
|
NM_015266
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr16_-_4243762
|
0.414
|
|
LOC100507501
|
hypothetical LOC100507501
|
|
chr1_-_153799186
|
0.414
|
|
ASH1L
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr1_-_23376860
|
0.414
|
|
LUZP1
|
leucine zipper protein 1
|
|
chr14_-_35859545
|
0.409
|
NM_001144891
NM_016586
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr10_+_124124196
|
0.408
|
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr11_-_1026654
|
0.397
|
NM_005961
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming
|
|
chr19_-_47438518
|
0.393
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr2_-_151826504
|
0.392
|
NM_198557
|
RBM43
|
RNA binding motif protein 43
|
|
chr3_-_46012303
|
0.391
|
NM_024513
|
FYCO1
|
FYVE and coiled-coil domain containing 1
|
|
chr6_-_48186868
|
0.389
|
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr16_+_2593351
|
0.386
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr1_+_117711575
|
0.382
|
NM_006699
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2
|
|
chr10_+_11824361
|
0.377
|
NM_024693
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|
|
chr4_+_153676857
|
0.376
|
|
DKFZP434I0714
|
hypothetical protein DKFZP434I0714
|
|
chr20_-_47238082
|
0.371
|
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr19_-_47438548
|
0.368
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr1_-_52936602
|
0.367
|
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr5_-_167938875
|
0.362
|
|
PANK3
|
pantothenate kinase 3
|
|
chr21_+_43186428
|
0.359
|
NM_001001503
NM_021075
|
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa
|
|
chr1_-_10455190
|
0.358
|
NM_004401
NM_213566
|
DFFA
|
DNA fragmentation factor, 45kDa, alpha polypeptide
|
|
chr8_-_22982435
|
0.357
|
NM_003842
NM_147187
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b
|
|
chr6_+_76515608
|
0.357
|
NM_004999
|
MYO6
|
myosin VI
|
|
chr11_+_1897374
|
0.353
|
NM_001042780
NM_001042781
NM_001042782
NM_006757
|
TNNT3
|
troponin T type 3 (skeletal, fast)
|
|
chr17_-_975809
|
0.348
|
|
ABR
|
active BCR-related gene
|
|
chr18_+_19523526
|
0.340
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr11_-_66993211
|
0.340
|
NM_001078650
NM_001078651
NM_025124
|
TMEM134
|
transmembrane protein 134
|
|
chr6_-_30818198
|
0.336
|
NM_005803
|
FLOT1
|
flotillin 1
|
|
chr16_-_74024750
|
0.334
|
NM_006324
|
CFDP1
|
craniofacial development protein 1
|
|
chr13_-_100866706
|
0.333
|
NM_052867
|
NALCN
|
sodium leak channel, non-selective
|
|
chr6_+_33486750
|
0.333
|
NM_002636
NM_024165
|
PHF1
|
PHD finger protein 1
|
|
chr9_+_103200939
|
0.331
|
NM_003452
NM_197977
|
ZNF189
|
zinc finger protein 189
|
|
chr20_-_5048443
|
0.331
|
NM_182649
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr20_-_47238310
|
0.331
|
NM_001037328
NM_004602
NM_017452
NM_017453
NM_017454
|
STAU1
|
staufen, RNA binding protein, homolog 1 (Drosophila)
|
|
chr10_+_47216925
|
0.323
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr14_-_22693333
|
0.321
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter, L-type), member 8
|
|
chr3_+_16901455
|
0.319
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr19_+_61344341
|
0.310
|
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr1_-_52936611
|
0.309
|
NM_023077
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr11_+_118544625
|
0.306
|
NM_024618
NM_170722
|
NLRX1
|
NLR family member X1
|
|
chr3_+_32123004
|
0.304
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr10_+_88404285
|
0.302
|
NM_001030015
NM_033282
|
OPN4
|
opsin 4
|
|
chr7_+_115953582
|
0.297
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr2_-_175741096
|
0.296
|
NM_001880
|
ATF2
|
activating transcription factor 2
|
|
chr21_-_42172476
|
0.291
|
NM_001040424
NM_022115
|
PRDM15
|
PR domain containing 15
|
|
chr1_+_42920946
|
0.289
|
|
YBX1
|
Y box binding protein 1
|
|
chr12_+_47498778
|
0.288
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr16_-_87244896
|
0.287
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr1_-_6375949
|
0.283
|
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr17_+_20423628
|
0.281
|
NM_001190790
|
CDRT15L2
|
CMT1A duplicated region transcript 15-like 2
|
|
chr12_+_27377034
|
0.280
|
NM_020183
|
ARNTL2
|
aryl hydrocarbon receptor nuclear translocator-like 2
|
|
chr16_-_30841938
|
0.277
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr20_+_4077409
|
0.277
|
NM_175839
NM_175840
NM_175841
NM_175842
|
SMOX
|
spermine oxidase
|
|
chr3_-_184363126
|
0.276
|
|
LAMP3
|
lysosomal-associated membrane protein 3
|
|
chr19_+_1358664
|
0.269
|
|
DAZAP1
|
DAZ associated protein 1
|
|
chr8_-_57286412
|
0.268
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr11_+_3833495
|
0.266
|
NM_003156
|
STIM1
|
stromal interaction molecule 1
|
|
chr3_-_44494108
|
0.262
|
|
ZNF445
|
zinc finger protein 445
|
|
chr19_-_43406621
|
0.262
|
NM_001135155
NM_004647
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr19_+_40183169
|
0.262
|
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr22_+_21852551
|
0.261
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr1_+_11718798
|
0.261
|
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr19_+_13090101
|
0.260
|
NM_052876
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr1_-_201202956
|
0.260
|
NM_016243
|
CYB5R1
|
cytochrome b5 reductase 1
|
|
chr2_+_197212522
|
0.257
|
NM_001080539
|
CCDC150
|
coiled-coil domain containing 150
|
|
chr17_-_40122542
|
0.255
|
NM_001099225
NM_144609
|
CCDC43
|
coiled-coil domain containing 43
|
|
chr17_-_14080874
|
0.254
|
NM_001007530
|
CDRT15
|
CMT1A duplicated region transcript 15
|
|
chr2_+_11213053
|
0.252
|
|
PQLC3
|
PQ loop repeat containing 3
|
|
chr6_+_146177723
|
0.249
|
|
LOC100507557
|
hypothetical LOC100507557
|
|
chr17_-_9870292
|
0.249
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr8_+_95801375
|
0.249
|
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr8_-_23077392
|
0.247
|
NM_003840
|
TNFRSF10D
|
tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain
|
|
chr2_+_176761552
|
0.247
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr7_+_43589163
|
0.246
|
NM_004760
|
STK17A
|
serine/threonine kinase 17a
|
|
chr19_-_52426055
|
0.246
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr3_-_44494160
|
0.245
|
NM_181489
|
ZNF445
|
zinc finger protein 445
|
|
chr5_-_137099361
|
0.243
|
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr1_+_232107170
|
0.243
|
NM_173508
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr8_+_95801277
|
0.240
|
NM_181787
|
DPY19L4
|
dpy-19-like 4 (C. elegans)
|
|
chr3_+_9919511
|
0.239
|
NM_001193380
NM_153480
|
IL17RE
|
interleukin 17 receptor E
|
|
chrX_+_153767803
|
0.239
|
NM_001007523
NM_001007524
NM_012151
|
F8A2
F8A3
F8A1
|
coagulation factor VIII-associated (intronic transcript) 2
coagulation factor VIII-associated (intronic transcript) 3
coagulation factor VIII-associated (intronic transcript) 1
|
|
chr22_-_22422951
|
0.238
|
NM_021916
|
ZNF70
|
zinc finger protein 70
|
|
chr9_+_88953378
|
0.237
|
NM_001001709
|
C9orf170
|
chromosome 9 open reading frame 170
|
|
chr5_-_167938991
|
0.237
|
NM_024594
|
PANK3
|
pantothenate kinase 3
|
|
chr11_+_85017269
|
0.234
|
NM_001193537
NM_001193538
NM_018480
|
TMEM126B
|
transmembrane protein 126B
|
|
chr20_+_56900116
|
0.231
|
NM_001077488
|
GNAS
|
GNAS complex locus
|
|
chr2_+_97717300
|
0.231
|
NM_207519
|
ZAP70
|
zeta-chain (TCR) associated protein kinase 70kDa
|
|
chr6_+_37895570
|
0.231
|
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr20_+_4077466
|
0.230
|
|
SMOX
|
spermine oxidase
|
|
chr2_+_113546365
|
0.230
|
NM_032556
|
IL1F10
|
interleukin 1 family, member 10 (theta)
|
|
chr12_-_74710741
|
0.230
|
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr15_-_40173949
|
0.224
|
NM_178034
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic)
|
|
chr6_+_76515690
|
0.221
|
|
MYO6
|
myosin VI
|
|
chrX_+_129133234
|
0.221
|
NM_004794
|
RAB33A
|
RAB33A, member RAS oncogene family
|
|
chr1_+_36169260
|
0.220
|
NM_024852
NM_177422
|
EIF2C3
|
eukaryotic translation initiation factor 2C, 3
|
|
chr2_+_171998988
|
0.220
|
NM_001164821
NM_025000
|
DCAF17
|
DDB1 and CUL4 associated factor 17
|
|
chr16_-_1410710
|
0.219
|
|
C16orf91
|
chromosome 16 open reading frame 91
|
|
chr6_-_30820264
|
0.219
|
NM_003897
|
IER3
|
immediate early response 3
|
|
chr7_+_65307691
|
0.218
|
NM_003596
|
TPST1
|
tyrosylprotein sulfotransferase 1
|
|
chr11_+_63758550
|
0.217
|
NM_003377
|
VEGFB
|
vascular endothelial growth factor B
|
|
chr16_-_29781842
|
0.216
|
NM_006319
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase
|
|
chr21_+_45649429
|
0.215
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr13_-_100866839
|
0.215
|
|
NALCN
|
sodium leak channel, non-selective
|
|
chr3_+_32123140
|
0.214
|
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr22_-_21231421
|
0.213
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr4_-_108860740
|
0.212
|
NM_005443
|
PAPSS1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1
|
|
chr2_-_24161033
|
0.211
|
NM_147184
NM_004881
|
TP53I3
|
tumor protein p53 inducible protein 3
|
|
chr16_-_56789142
|
0.211
|
NM_001896
|
CSNK2A2
|
casein kinase 2, alpha prime polypeptide
|
|
chr7_+_100557098
|
0.209
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chrX_+_38548316
|
0.208
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr20_+_61760084
|
0.208
|
NM_016434
NM_032957
|
RTEL1
|
regulator of telomere elongation helicase 1
|
|
chrX_-_153767625
|
0.206
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr10_-_95231933
|
0.205
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr22_-_17846621
|
0.205
|
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr6_+_21774624
|
0.204
|
|
FLJ22536
|
hypothetical locus LOC401237
|
|
chr4_-_159863889
|
0.202
|
NM_005038
|
PPID
|
peptidylprolyl isomerase D
|
|
chr15_+_83724743
|
0.201
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr12_+_30970882
|
0.200
|
|
TSPAN11
|
tetraspanin 11
|
|
chr20_-_33007242
|
0.199
|
|
GSS
|
glutathione synthetase
|
|
chr1_+_11718715
|
0.198
|
NM_001040194
NM_001040195
NM_001040196
NM_001040197
NM_020350
|
AGTRAP
|
angiotensin II receptor-associated protein
|
|
chr20_+_34667624
|
0.196
|
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr9_-_5997891
|
0.196
|
NM_001017969
|
KIAA2026
|
KIAA2026
|
|
chr20_-_33007243
|
0.196
|
NM_000178
|
GSS
|
glutathione synthetase
|
|
chr20_+_56899904
|
0.193
|
|
GNAS
|
GNAS complex locus
|
|
chr22_+_39096509
|
0.193
|
NM_015705
|
SGSM3
|
small G protein signaling modulator 3
|
|
chr10_+_124124083
|
0.192
|
NM_001001974
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr20_-_60435983
|
0.191
|
NM_080833
|
C20orf151
|
chromosome 20 open reading frame 151
|
|
chr19_+_55985483
|
0.190
|
NM_033068
|
ACPT
|
acid phosphatase, testicular
|
|
chrX_+_151837064
|
0.190
|
NM_001178113
|
ZNF185
|
zinc finger protein 185 (LIM domain)
|
|
chr20_-_33007209
|
0.188
|
|
GSS
|
glutathione synthetase
|
|
chr19_+_32976717
|
0.187
|
|
|
|
|
chr7_-_44496703
|
0.187
|
NM_015332
|
NUDCD3
|
NudC domain containing 3
|
|
chr1_+_54954082
|
0.183
|
NM_004623
|
TTC4
|
tetratricopeptide repeat domain 4
|
|
chr4_-_184662604
|
0.181
|
|
LOC389247
|
hypothetical LOC389247
|
|
chrX_+_38548373
|
0.181
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr17_-_58877276
|
0.179
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr2_-_69724480
|
0.179
|
NM_014911
|
AAK1
|
AP2 associated kinase 1
|
|
chr3_+_161601040
|
0.178
|
NM_005496
|
SMC4
|
structural maintenance of chromosomes 4
|
|
chr17_-_39447817
|
0.178
|
NM_032376
|
TMEM101
|
transmembrane protein 101
|
|
chr2_+_187266899
|
0.177
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr19_-_14089362
|
0.176
|
NM_002730
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha
|
|
chr19_+_47056162
|
0.176
|
|
RPS19
|
ribosomal protein S19
|
|
chr18_+_3584056
|
0.176
|
|
FLJ35776
|
hypothetical LOC649446
|
|
chr1_-_207892420
|
0.176
|
NM_000228
|
LAMB3
|
laminin, beta 3
|
|
chr14_+_101853466
|
0.175
|
|
ZNF839
|
zinc finger protein 839
|
|
chr2_-_105312536
|
0.175
|
NM_001142621
NM_004257
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1
|