|
chr14_-_91576037
|
1.337
|
NM_004239
|
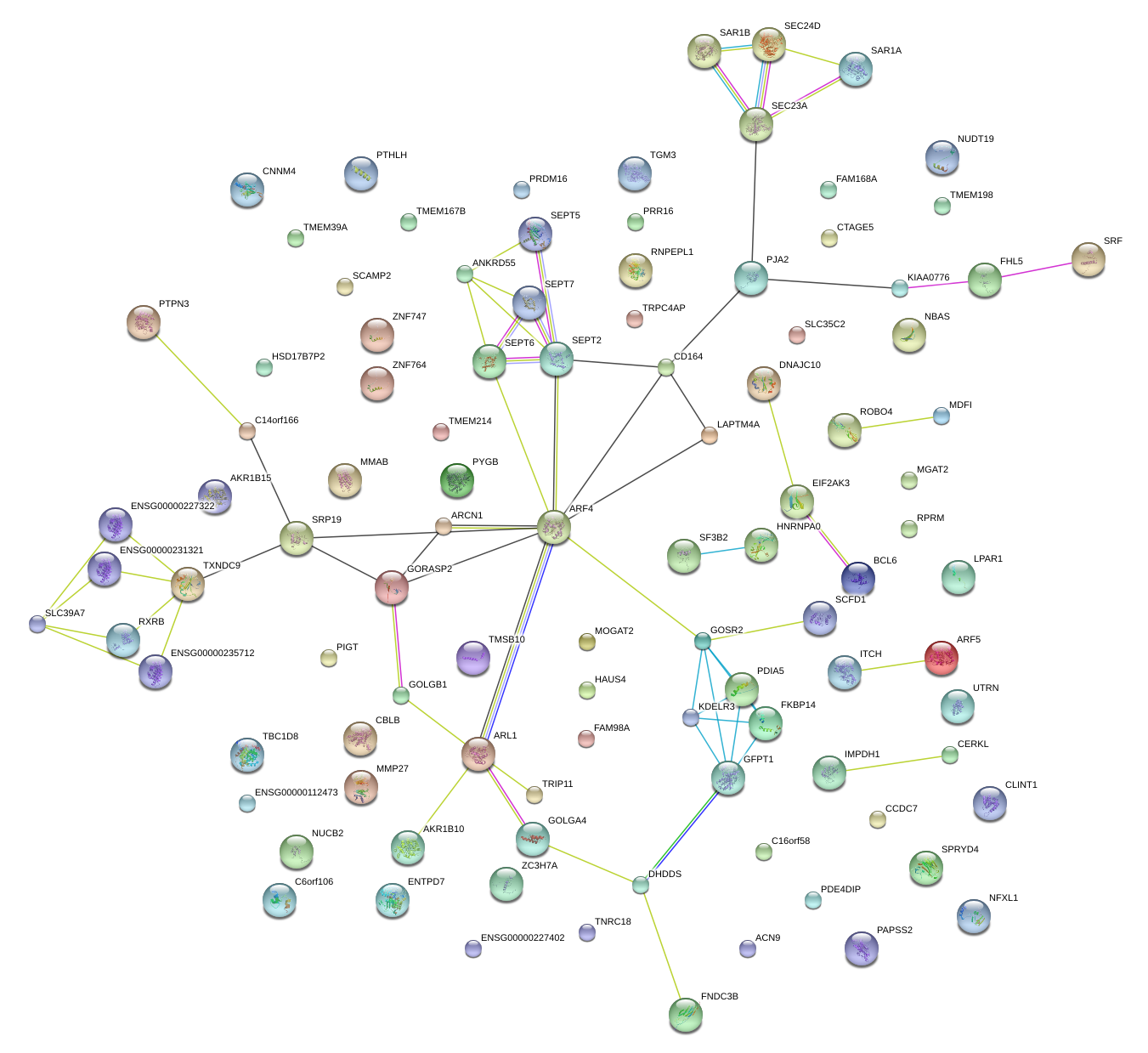
TRIP11
|
thyroid hormone receptor interactor 11
|
|
chr1_+_26631359
|
1.302
|
NM_024887
NM_205861
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr10_+_89409332
|
1.269
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr2_+_183289205
|
1.184
|
NM_018981
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10
|
|
chr20_+_43478120
|
1.146
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr6_+_144649142
|
1.131
|
|
UTRN
|
utrophin
|
|
chr15_-_72952648
|
1.121
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr14_-_38642165
|
1.067
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr14_+_30161267
|
1.059
|
NM_016106
NM_182835
|
SCFD1
|
sec1 family domain containing 1
|
|
chr14_-_38642029
|
1.058
|
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr2_+_96790298
|
1.050
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr2_+_171494026
|
1.041
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr3_-_120665113
|
0.941
|
NM_018266
|
TMEM39A
|
transmembrane protein 39A
|
|
chr22_+_37193961
|
0.936
|
NM_006855
NM_016657
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr2_-_88707978
|
0.903
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr2_-_33677798
|
0.894
|
NM_015475
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr20_-_33144013
|
0.889
|
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr20_-_44426407
|
0.884
|
NM_015945
NM_173073
NM_173179
|
SLC35C2
|
solute carrier family 35, member C2
|
|
chr11_+_17254854
|
0.878
|
NM_005013
|
NUCB2
|
nucleobindin 2
|
|
chr1_-_143750988
|
0.861
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_-_69467825
|
0.837
|
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr5_+_112224875
|
0.824
|
NM_003135
|
SRP19
|
signal recognition particle 19kDa
|
|
chr14_+_38806280
|
0.813
|
|
CTAGE5
|
CTAGE family, member 5
|
|
chr2_+_84986273
|
0.770
|
NM_021103
|
TMSB10
|
thymosin beta 10
|
|
chr4_-_47611255
|
0.761
|
NM_152995
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1
|
|
chr3_+_173240983
|
0.754
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr19_+_37874529
|
0.738
|
NM_001105570
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19
|
|
chr20_+_25176698
|
0.701
|
NM_002862
|
PYGB
|
phosphorylase, glycogen; brain
|
|
chr5_-_108773468
|
0.696
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr12_-_28014151
|
0.682
|
NM_198964
NM_198966
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr12_+_55148567
|
0.681
|
NM_207344
|
SPRYD4
|
SPRY domain containing 4
|
|
chr2_-_20115269
|
0.660
|
NM_014713
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr20_+_32414715
|
0.655
|
NM_031483
|
ITCH
|
itchy E3 ubiquitin protein ligase homolog (mouse)
|
|
chr20_-_33144254
|
0.645
|
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr20_-_33144262
|
0.644
|
NM_015638
NM_199368
|
TRPC4AP
|
transient receptor potential cation channel, subfamily C, member 4 associated protein
|
|
chr5_+_119827903
|
0.631
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr16_-_30477006
|
0.631
|
NM_001172679
NM_033410
|
ZNF764
|
zinc finger protein 764
|
|
chr14_+_38806028
|
0.622
|
NM_005930
NM_203355
|
CTAGE5
|
CTAGE family, member 5
|
|
chr16_-_11798507
|
0.615
|
|
ZC3H7A
|
zinc finger CCCH-type containing 7A
|
|
chr2_+_171493870
|
0.614
|
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr12_-_100325630
|
0.613
|
NM_001177
|
ARL1
|
ADP-ribosylation factor-like 1
|
|
chr3_-_57557906
|
0.609
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr6_+_41714171
|
0.596
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr1_-_143643324
|
0.595
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr7_-_5428043
|
0.568
|
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr3_+_124268545
|
0.567
|
NM_006810
|
PDIA5
|
protein disulfide isomerase family A, member 5
|
|
chr7_-_30032649
|
0.563
|
NM_017946
|
FKBP14
|
FK506 binding protein 14, 22 kDa
|
|
chr2_-_101134146
|
0.550
|
NM_001102426
|
TBC1D8
|
TBC1 domain family, member 8 (with GRAM domain)
|
|
chr6_+_33276580
|
0.547
|
NM_001077516
NM_006979
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7
|
|
chr11_+_65576379
|
0.546
|
NM_006842
|
SF3B2
|
splicing factor 3b, subunit 2, 145kDa
|
|
chr4_-_119976631
|
0.525
|
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr6_+_97076390
|
0.524
|
NM_015323
|
KIAA0776
|
KIAA0776
|
|
chr3_-_107070400
|
0.518
|
NM_170662
|
CBLB
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence b
|
|
chr3_+_37259684
|
0.516
|
NM_001172713
NM_002078
|
GOLGA4
|
golgin A4
|
|
chr11_-_102081654
|
0.514
|
NM_022122
|
MMP27
|
matrix metallopeptidase 27
|
|
chr5_-_137116746
|
0.498
|
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr11_-_72986738
|
0.488
|
NM_015159
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr10_+_32775027
|
0.476
|
NM_001026383
NM_145023
|
CCDC7
|
coiled-coil domain containing 7
|
|
chr2_-_15618865
|
0.469
|
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr17_+_42355492
|
0.465
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr14_-_22904074
|
0.454
|
|
|
|
|
chr1_+_109434896
|
0.450
|
NM_020141
|
TMEM167B
|
transmembrane protein 167B
|
|
chr6_+_97076451
|
0.449
|
|
KIAA0776
|
KIAA0776
|
|
chr14_-_22496098
|
0.445
|
NM_001166269
NM_001166270
NM_017815
|
HAUS4
|
HAUS augmin-like complex, subunit 4
|
|
chr6_-_33276416
|
0.437
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr2_+_241156776
|
0.435
|
NM_018226
|
RNPEPL1
|
arginyl aminopeptidase (aminopeptidase B)-like 1
|
|
chr2_-_182229977
|
0.434
|
NM_001030311
NM_001030312
NM_001030313
NM_001160277
NM_201548
|
CERKL
|
ceramide kinase-like
|
|
chr9_-_112840064
|
0.433
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr2_-_15618896
|
0.433
|
NM_015909
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr11_+_117948350
|
0.430
|
|
ARCN1
|
archain 1
|
|
chr9_-_111300350
|
0.428
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr2_+_84986510
|
0.426
|
|
TMSB10
|
thymosin beta 10
|
|
chr6_-_33276426
|
0.426
|
|
RXRB
|
retinoid X receptor, beta
|
|
chr14_+_30161285
|
0.418
|
|
SCFD1
|
sec1 family domain containing 1
|
|
chr2_-_20114761
|
0.417
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr10_-_71600191
|
0.415
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr10_+_101409159
|
0.415
|
NM_020354
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr16_-_31427130
|
0.404
|
NM_022744
|
C16orf58
|
chromosome 16 open reading frame 58
|
|
chr15_-_61680003
|
0.402
|
|
LOC100130855
|
hypothetical LOC100130855
|
|
chr3_-_122951200
|
0.400
|
NM_004487
|
GOLGB1
|
golgin B1
|
|
chr2_+_241903395
|
0.391
|
NM_001008491
NM_006155
|
SEPT2
|
septin 2
|
|
chr7_-_127833231
|
0.390
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr3_-_188945921
|
0.379
|
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr11_+_117948301
|
0.373
|
NM_001142281
NM_001655
|
ARCN1
|
archain 1
|
|
chr7_+_96584929
|
0.368
|
|
ACN9
|
ACN9 homolog (S. cerevisiae)
|
|
chr14_+_51526071
|
0.361
|
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr16_-_30453499
|
0.357
|
|
ZNF747
|
zinc finger protein 747
|
|
chr2_+_220116986
|
0.351
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr2_+_27109341
|
0.349
|
|
TMEM214
|
transmembrane protein 214
|
|
chr14_+_51525942
|
0.346
|
NM_016039
|
C14orf166
|
chromosome 14 open reading frame 166
|
|
chr5_-_157218555
|
0.345
|
NM_001195555
NM_001195556
NM_014666
|
CLINT1
|
clathrin interactor 1
|
|
chr6_-_109809595
|
0.344
|
|
CD164
|
CD164 molecule, sialomucin
|
|
chr6_-_34772495
|
0.343
|
|
C6orf106
|
chromosome 6 open reading frame 106
|
|
chr6_-_86445138
|
0.341
|
|
SNHG5
|
small nucleolar RNA host gene 5 (non-protein coding)
|
|
chr14_+_49157150
|
0.339
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr2_-_154043403
|
0.337
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr2_-_99319197
|
0.331
|
NM_005783
|
TXNDC9
|
thioredoxin domain containing 9
|
|
chr5_+_126881207
|
0.330
|
NM_130809
|
PRRC1
|
proline-rich coiled-coil 1
|
|
chr3_-_73756668
|
0.329
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr15_-_49817454
|
0.326
|
NM_153374
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2
|
|
chr11_-_118477739
|
0.325
|
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase)
|
|
chr14_-_23734598
|
0.318
|
NM_001014842
NM_006405
|
TM9SF1
|
transmembrane 9 superfamily member 1
|
|
chr6_+_43036442
|
0.314
|
NM_018960
|
GNMT
|
glycine N-methyltransferase
|
|
chr2_+_220117009
|
0.312
|
|
TMEM198
|
transmembrane protein 198
|
|
chr10_+_104667991
|
0.312
|
NM_017649
NM_199076
NM_199077
|
CNNM2
|
cyclin M2
|
|
chr2_-_20114881
|
0.311
|
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha
|
|
chr1_-_206484258
|
0.308
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr8_+_95722516
|
0.306
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr10_+_106103503
|
0.302
|
NM_001008723
|
CCDC147
|
coiled-coil domain containing 147
|
|
chr12_+_61146798
|
0.295
|
NM_015026
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr6_-_34772565
|
0.291
|
NM_022758
NM_024294
|
C6orf106
|
chromosome 6 open reading frame 106
|
|
chr6_+_27208763
|
0.282
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr5_+_119827797
|
0.280
|
|
PRR16
|
proline rich 16
|
|
chr2_+_64604940
|
0.280
|
NM_001002243
NM_017657
NM_203437
|
AFTPH
|
aftiphilin
|
|
chr12_+_61147083
|
0.280
|
|
MON2
|
MON2 homolog (S. cerevisiae)
|
|
chr4_-_119976736
|
0.278
|
NM_014822
|
SEC24D
|
SEC24 family, member D (S. cerevisiae)
|
|
chr1_-_87152372
|
0.276
|
|
SEP15
|
15 kDa selenoprotein
|
|
chr2_+_200528173
|
0.274
|
NM_024520
|
C2orf47
|
chromosome 2 open reading frame 47
|
|
chr13_+_24940965
|
0.274
|
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr12_+_70519753
|
0.269
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr19_-_44214707
|
0.268
|
|
|
|
|
chr2_-_220116508
|
0.266
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr11_-_61441536
|
0.262
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr17_-_37828643
|
0.261
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr11_-_74819165
|
0.259
|
NM_001039548
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr17_+_42355482
|
0.258
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr15_-_81744469
|
0.256
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr11_+_113776582
|
0.254
|
NM_016090
|
RBM7
|
RNA binding motif protein 7
|
|
chr6_-_33276347
|
0.254
|
NM_021976
|
RXRB
|
retinoid X receptor, beta
|
|
chr10_-_22332368
|
0.252
|
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr9_-_129573330
|
0.248
|
NM_005489
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr2_+_203811903
|
0.247
|
|
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1
|
|
chr18_-_42956645
|
0.245
|
|
IER3IP1
|
immediate early response 3 interacting protein 1
|
|
chr12_+_67489063
|
0.242
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr1_+_159390379
|
0.239
|
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1
|
|
chr6_-_7855277
|
0.234
|
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr2_-_27147995
|
0.234
|
NM_001134693
|
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae)
|
|
chr17_+_3993690
|
0.232
|
|
CYB5D2
|
cytochrome b5 domain containing 2
|
|
chr19_+_51796399
|
0.229
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr2_+_220116596
|
0.228
|
|
TMEM198
|
transmembrane protein 198
|
|
chr1_-_87152396
|
0.227
|
|
SEP15
|
15 kDa selenoprotein
|
|
chr2_-_27195276
|
0.224
|
NM_001166239
|
CGREF1
|
cell growth regulator with EF-hand domain 1
|
|
chr13_+_24940917
|
0.223
|
|
ATP8A2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2
|
|
chr6_-_108386102
|
0.223
|
NM_007214
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr2_-_74915722
|
0.221
|
|
|
|
|
chr12_-_56445653
|
0.220
|
|
CYP27B1
|
cytochrome P450, family 27, subfamily B, polypeptide 1
|
|
chr19_-_54636425
|
0.220
|
NM_020309
|
SLC17A7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7
|
|
chr6_+_1336077
|
0.217
|
|
|
|
|
chr16_+_51722285
|
0.215
|
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr22_+_44828338
|
0.213
|
|
|
|
|
chr18_-_42956733
|
0.206
|
NM_016097
|
IER3IP1
|
immediate early response 3 interacting protein 1
|
|
chr14_+_49135164
|
0.204
|
NM_152329
NM_203467
|
LRR1
|
leucine rich repeat protein 1
|
|
chr3_+_133861894
|
0.202
|
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr12_+_58276114
|
0.201
|
|
SLC16A7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2)
|
|
chr11_+_113776504
|
0.200
|
|
RBM7
|
RNA binding motif protein 7
|
|
chr2_+_27109249
|
0.200
|
NM_001083590
NM_017727
|
TMEM214
|
transmembrane protein 214
|
|
chr5_-_121441805
|
0.199
|
|
LOX
|
lysyl oxidase
|
|
chr22_-_29015452
|
0.198
|
NM_001037666
|
GATSL3
|
GATS protein-like 3
|
|
chr3_+_171167243
|
0.197
|
NM_003262
|
SEC62
|
SEC62 homolog (S. cerevisiae)
|
|
chr3_-_134863316
|
0.196
|
NM_007027
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1
|
|
chrX_+_152565849
|
0.195
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr3_+_129253997
|
0.195
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr3_-_57558120
|
0.194
|
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr4_-_122063118
|
0.192
|
NM_018699
|
PRDM5
|
PR domain containing 5
|
|
chr2_+_27740254
|
0.191
|
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein
|
|
chr2_+_202607554
|
0.190
|
NM_003507
|
FZD7
|
frizzled homolog 7 (Drosophila)
|
|
chr2_+_64605049
|
0.189
|
|
AFTPH
|
aftiphilin
|
|
chr3_+_101462375
|
0.186
|
NM_018309
|
TBC1D23
|
TBC1 domain family, member 23
|
|
chr17_+_52688842
|
0.184
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr3_+_129253977
|
0.183
|
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr4_-_39136448
|
0.182
|
NM_001024921
|
RPL9
|
ribosomal protein L9
|
|
chr6_-_108385905
|
0.182
|
|
SEC63
|
SEC63 homolog (S. cerevisiae)
|
|
chr18_-_42956730
|
0.181
|
|
IER3IP1
|
immediate early response 3 interacting protein 1
|
|
chr3_+_129254032
|
0.181
|
|
|
|
|
chr6_-_7855125
|
0.177
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr2_+_75039273
|
0.177
|
NM_019896
|
POLE4
|
polymerase (DNA-directed), epsilon 4 (p12 subunit)
|
|
chr1_+_234625338
|
0.175
|
NM_080738
|
EDARADD
|
EDAR-associated death domain
|
|
chr2_-_71210856
|
0.175
|
NM_032601
|
MCEE
|
methylmalonyl CoA epimerase
|
|
chr12_+_88268845
|
0.172
|
|
LOC100131490
|
hypothetical LOC100131490
|
|
chr3_+_129253816
|
0.171
|
NM_013336
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae)
|
|
chr8_+_120289735
|
0.171
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr9_+_91115924
|
0.170
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr17_+_14145051
|
0.168
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr17_+_18625243
|
0.166
|
NM_016078
|
FAM18B1
|
family with sequence similarity 18, member B1
|
|
chr19_+_47080285
|
0.165
|
NM_199002
|
ARHGEF1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr6_-_7855715
|
0.165
|
NM_030810
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr10_+_92970487
|
0.163
|
|
PCGF5
|
polycomb group ring finger 5
|
|
chr20_+_49008754
|
0.162
|
NM_014484
|
MOCS3
|
molybdenum cofactor synthesis 3
|
|
chr5_-_121441910
|
0.162
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr2_+_27109327
|
0.162
|
|
TMEM214
|
transmembrane protein 214
|
|
chr16_-_57276108
|
0.160
|
NM_018231
|
SLC38A7
|
solute carrier family 38, member 7
|
|
chr11_+_124967961
|
0.159
|
|
STT3A
|
STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
|
|
chr3_-_180651874
|
0.158
|
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr20_+_61054539
|
0.158
|
|
SLC17A9
|
solute carrier family 17, member 9
|
|
chr14_-_49157076
|
0.157
|
NM_001001
|
RPL36AL
|
ribosomal protein L36a-like
|
|
chr14_-_22904218
|
0.157
|
|
EFS
|
embryonal Fyn-associated substrate
|
|
chr11_+_61647990
|
0.157
|
NM_001040694
NM_020238
|
INCENP
|
inner centromere protein antigens 135/155kDa
|
|
chr14_+_38805622
|
0.156
|
|
CTAGE5
|
CTAGE family, member 5
|
|
chr1_-_21982037
|
0.154
|
|
USP48
|
ubiquitin specific peptidase 48
|
|
chr11_-_65813031
|
0.154
|
|
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae)
|