|
chr3_+_35696119
|
2.124
|
NM_001025068
NM_001025069
|
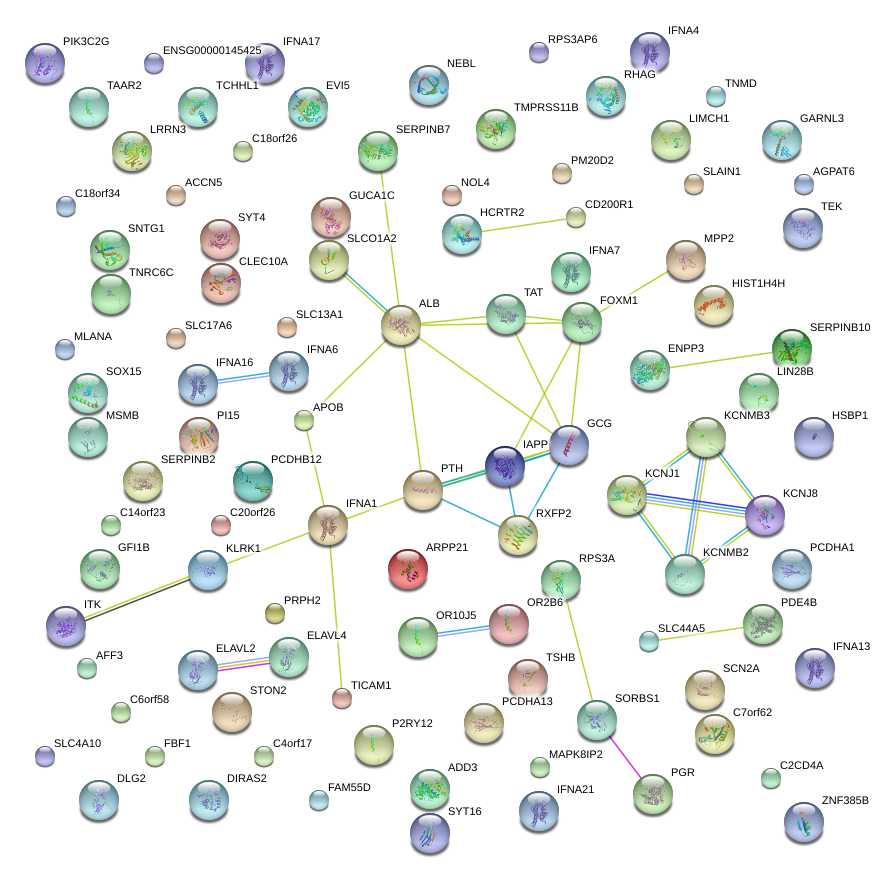
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr14_+_28311740
|
2.042
|
|
C14orf23
|
chromosome 14 open reading frame 23
|
|
chr16_-_70168449
|
1.902
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr11_-_13474101
|
1.590
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr4_-_149577404
|
1.431
|
|
|
|
|
chr18_+_59593591
|
1.389
|
NM_003784
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7
|
|
chr18_-_29882555
|
1.349
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr4_+_41309690
|
1.265
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr18_-_39111243
|
1.253
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr6_+_127940011
|
1.235
|
NM_001010905
|
C6orf58
|
chromosome 6 open reading frame 58
|
|
chr6_+_132000134
|
1.221
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr2_-_162717159
|
1.195
|
NM_002054
|
GCG
|
glucagon
|
|
chr10_+_111755715
|
1.189
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr6_+_55147029
|
1.163
|
NM_001526
|
HCRTR2
|
hypocretin (orexin) receptor 2
|
|
chr4_+_41309668
|
1.132
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr2_+_162189090
|
1.116
|
NM_001178015
NM_001178016
NM_022058
|
SLC4A10
|
solute carrier family 4, sodium bicarbonate transporter, member 10
|
|
chr12_+_21417068
|
1.062
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr4_+_41309482
|
1.038
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr15_+_60146467
|
1.011
|
NM_207322
|
C2CD4A
|
C2 calcium-dependent domain containing 4A
|
|
chr8_+_75899326
|
0.996
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr14_-_80934523
|
0.988
|
NM_033104
|
STON2
|
stonin 2
|
|
chr9_+_129066576
|
0.973
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr10_-_21226536
|
0.943
|
NM_006393
|
NEBL
|
nebulette
|
|
chr2_+_165804157
|
0.901
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr7_-_122627235
|
0.862
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chrX_+_99726445
|
0.853
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr9_+_134843918
|
0.853
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr11_-_113971607
|
0.849
|
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr11_-_100505667
|
0.848
|
NM_000926
|
PGR
|
progesterone receptor
|
|
chr11_-_128242342
|
0.833
|
NM_153764
NM_153765
NM_153766
NM_153767
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr9_+_5880801
|
0.814
|
NM_005511
|
MLANA
|
melan-A
|
|
chr12_-_21379098
|
0.789
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr9_-_21207303
|
0.786
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr6_-_49712483
|
0.763
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr14_+_61532293
|
0.760
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr4_+_100651183
|
0.752
|
NM_032149
|
C4orf17
|
chromosome 4 open reading frame 17
|
|
chr5_+_140568451
|
0.738
|
NM_018932
|
PCDHB12
|
protocadherin beta 12
|
|
chr9_-_21177529
|
0.726
|
NM_021068
|
IFNA4
|
interferon, alpha 4
|
|
chr12_-_21818881
|
0.713
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr15_-_41195994
|
0.696
|
|
RPS3A
|
ribosomal protein S3A
|
|
chr1_-_93030538
|
0.696
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr6_+_105511615
|
0.688
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr18_+_50409387
|
0.687
|
NM_173629
|
C18orf26
|
chromosome 18 open reading frame 26
|
|
chr11_-_113971681
|
0.664
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr18_+_59733724
|
0.662
|
NM_005024
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr11_-_128217550
|
0.650
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr7_-_88262966
|
0.650
|
NM_152706
|
C7orf62
|
chromosome 7 open reading frame 62
|
|
chr2_-_180319013
|
0.618
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chr5_+_156540425
|
0.617
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr1_-_150328153
|
0.606
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr1_+_50344550
|
0.598
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr17_-_7434113
|
0.597
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr12_+_18305740
|
0.585
|
NM_004570
|
PIK3C2G
|
phosphoinositide-3-kinase, class 2, gamma polypeptide
|
|
chr9_-_21358005
|
0.574
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|
|
chr13_+_77213295
|
0.569
|
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr9_-_92444886
|
0.564
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr4_-_68793928
|
0.550
|
NM_182502
|
TMPRSS11B
|
transmembrane protease, serine 11B
|
|
chr1_+_66230977
|
0.550
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr1_-_75849350
|
0.542
|
NM_001130058
NM_152697
|
SLC44A5
|
solute carrier family 44, member 5
|
|
chr18_-_29273979
|
0.527
|
NM_001105528
|
C18orf34
|
chromosome 18 open reading frame 34
|
|
chr5_+_140146059
|
0.526
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr20_+_19985289
|
0.510
|
NM_001167816
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr6_-_132980901
|
0.495
|
NM_014626
|
TAAR2
|
trace amine associated receptor 2
|
|
chr3_-_110155237
|
0.492
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr3_-_152541129
|
0.491
|
NM_176876
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chr8_+_50987149
|
0.489
|
NM_018967
|
SNTG1
|
syntrophin, gamma 1
|
|
chr2_-_21106291
|
0.485
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr6_+_28032997
|
0.476
|
NM_012367
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6
|
|
chr4_+_74499651
|
0.475
|
|
ALB
|
albumin
|
|
chr2_-_100088760
|
0.474
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr22_+_49388427
|
0.471
|
NM_016431
|
MAPK8IP2
|
mitogen-activated protein kinase 8 interacting protein 2
|
|
chr10_+_51219558
|
0.470
|
NM_002443
NM_138634
|
MSMB
|
microseminoprotein, beta-
|
|
chr1_-_157772420
|
0.469
|
NM_001004469
|
OR10J5
|
olfactory receptor, family 10, subfamily J, member 5
|
|
chr9_+_27099138
|
0.468
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr11_+_22316242
|
0.467
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr7_+_110518297
|
0.467
|
NM_001099658
NM_001099660
NM_018334
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr4_-_157006827
|
0.464
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chr9_-_21340885
|
0.464
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr1_+_115373937
|
0.463
|
NM_000549
|
TSHB
|
thyroid stimulating hormone, beta
|
|
chr13_+_31211678
|
0.461
|
NM_001166058
NM_130806
|
RXFP2
|
relaxin/insulin-like family peptide receptor 2
|
|
chr11_-_83706027
|
0.458
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr10_-_97190885
|
0.456
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr12_-_10433852
|
0.452
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr17_+_73511843
|
0.450
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr3_+_179759181
|
0.449
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr11_-_84311867
|
0.447
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr12_+_56802
|
0.446
|
NM_015232
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr6_-_46155937
|
0.445
|
NM_001114086
|
CLIC5
|
chloride intracellular channel 5
|
|
chrX_-_15312390
|
0.443
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr7_+_18502342
|
0.439
|
NM_058176
NM_178425
|
HDAC9
|
histone deacetylase 9
|
|
chr15_+_70477707
|
0.434
|
NM_001080462
|
TMEM202
|
transmembrane protein 202
|
|
chr12_-_9123126
|
0.433
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr3_+_99484421
|
0.432
|
NM_001005482
|
OR5H2
|
olfactory receptor, family 5, subfamily H, member 2
|
|
chr4_-_80548378
|
0.432
|
NM_033214
|
GK2
|
glycerol kinase 2
|
|
chr4_+_47403107
|
0.428
|
|
|
|
|
chr11_-_84312085
|
0.421
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_-_156876798
|
0.417
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr20_-_13919256
|
0.416
|
NM_025229
|
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans)
|
|
chr18_-_30056431
|
0.416
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr9_-_21067804
|
0.416
|
NM_002176
|
IFNB1
|
interferon, beta 1, fibroblast
|
|
chr1_+_86707113
|
0.410
|
NM_001285
|
CLCA1
|
chloride channel accessory 1
|
|
chr1_-_214963279
|
0.409
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr3_+_103636548
|
0.404
|
NM_175056
|
ZPLD1
|
zona pellucida-like domain containing 1
|
|
chr6_-_71069347
|
0.402
|
NM_001851
|
COL9A1
|
collagen, type IX, alpha 1
|
|
chr4_+_91375204
|
0.399
|
NM_207491
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr8_-_7710547
|
0.398
|
NM_001037668
NM_001040705
|
DEFB107A
DEFB107B
|
defensin, beta 107A
defensin, beta 107B
|
|
chr3_+_191588354
|
0.391
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr6_+_52159143
|
0.388
|
NM_002190
|
IL17A
|
interleukin 17A
|
|
chr7_-_113346284
|
0.383
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A
|
|
chrX_-_83644054
|
0.382
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr2_+_214984705
|
0.375
|
NM_001080500
|
VWC2L
|
von Willebrand factor C domain containing protein 2-like
|
|
chr10_-_73518772
|
0.372
|
NM_001134434
NM_014767
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr1_+_156716290
|
0.371
|
NM_001004472
|
OR10R2
|
olfactory receptor, family 10, subfamily R, member 2
|
|
chr8_-_110762409
|
0.371
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr6_+_132901119
|
0.371
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr18_+_59374372
|
0.367
|
NM_080474
|
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12
|
|
chr14_-_105623358
|
0.364
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr20_-_19984685
|
0.362
|
NM_016652
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr4_+_74496660
|
0.361
|
|
ALB
|
albumin
|
|
chr11_+_55407348
|
0.358
|
NM_032681
|
SPRYD5
|
SPRY domain containing 5
|
|
chrX_-_106336199
|
0.357
|
NM_017681
|
NUP62CL
|
nucleoporin 62kDa C-terminal like
|
|
chr5_+_54434230
|
0.354
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr4_-_87500398
|
0.354
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr17_+_4434024
|
0.352
|
NM_198501
|
SMTNL2
|
smoothelin-like 2
|
|
chr4_-_77176163
|
0.346
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr2_-_207291364
|
0.346
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr11_+_110631916
|
0.345
|
NM_198498
|
C11orf53
|
chromosome 11 open reading frame 53
|
|
chr8_+_50147455
|
0.343
|
NM_001007176
|
C8orf22
|
chromosome 8 open reading frame 22
|
|
chr4_+_74496575
|
0.343
|
|
ALB
|
albumin
|
|
chr2_+_211166323
|
0.343
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr9_+_99303282
|
0.341
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr11_+_27019084
|
0.341
|
NM_003986
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1
|
|
chr11_+_22652957
|
0.339
|
|
GAS2
|
growth arrest-specific 2
|
|
chr3_+_134976965
|
0.339
|
|
TF
|
transferrin
|
|
chr4_+_119174947
|
0.335
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr4_-_123596921
|
0.334
|
NM_000586
|
IL2
|
interleukin 2
|
|
chr1_+_111818111
|
0.333
|
NM_174896
|
C1orf162
|
chromosome 1 open reading frame 162
|
|
chr10_+_90511142
|
0.333
|
NM_001102469
|
LIPN
|
lipase, family member N
|
|
chr6_-_32665409
|
0.333
|
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr11_+_22652894
|
0.331
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr2_+_204440753
|
0.327
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr6_+_127006298
|
0.325
|
|
LOC728666
|
PRELI domain containing 1 pseudogene
|
|
chr15_+_69626594
|
0.324
|
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr12_-_9252227
|
0.323
|
NM_002864
|
PZP
|
pregnancy-zone protein
|
|
chr1_+_150902495
|
0.322
|
NM_178430
|
LCE2D
|
late cornified envelope 2D
|
|
chr15_+_30721161
|
0.321
|
NM_001144757
NM_003020
|
SCG5
|
secretogranin V (7B2 protein)
|
|
chr20_+_9443426
|
0.320
|
|
C20orf103
|
chromosome 20 open reading frame 103
|
|
chr12_+_53534565
|
0.317
|
NM_058173
|
MUCL1
|
mucin-like 1
|
|
chrX_-_118610880
|
0.315
|
NM_001173488
|
NKRF
|
NFKB repressing factor
|
|
chr12_+_9871343
|
0.313
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chr11_-_27679617
|
0.312
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr6_+_132933153
|
0.311
|
NM_175067
|
TAAR6
|
trace amine associated receptor 6
|
|
chr3_-_148604760
|
0.307
|
NM_001168378
|
ZIC4
|
Zic family member 4
|
|
chr11_-_30562324
|
0.306
|
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr18_+_59705918
|
0.306
|
NM_001143818
NM_002575
|
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2
|
|
chr7_+_80145461
|
0.304
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr16_+_74868676
|
0.303
|
NM_033401
|
CNTNAP4
|
contactin associated protein-like 4
|
|
chr4_-_95482994
|
0.302
|
NM_014485
|
HPGDS
|
hematopoietic prostaglandin D synthase
|
|
chr7_+_142590633
|
0.294
|
NM_176881
|
TAS2R39
|
taste receptor, type 2, member 39
|
|
chr4_+_69465107
|
0.294
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr12_+_54080480
|
0.292
|
NM_001005518
|
OR6C65
|
olfactory receptor, family 6, subfamily C, member 65
|
|
chrX_+_106057984
|
0.291
|
|
CLDN2
|
claudin 2
|
|
chr7_+_29486010
|
0.289
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr4_+_14950657
|
0.289
|
NM_001135170
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr1_+_25471470
|
0.288
|
NM_001127691
NM_016124
|
RHD
|
Rh blood group, D antigen
|
|
chr7_-_16888121
|
0.287
|
NM_176813
|
AGR3
|
anterior gradient homolog 3 (Xenopus laevis)
|
|
chr12_-_16321885
|
0.286
|
NM_001170798
|
SLC15A5
|
solute carrier family 15, member 5
|
|
chr2_+_225973845
|
0.283
|
NM_020864
|
KIAA1486
|
KIAA1486
|
|
chr10_-_104587069
|
0.280
|
NM_000102
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1
|
|
chr10_-_69736600
|
0.276
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr6_-_133008834
|
0.275
|
NM_138327
|
TAAR1
|
trace amine associated receptor 1
|
|
chr11_+_123945102
|
0.273
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr3_-_131081910
|
0.269
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr12_+_8866416
|
0.268
|
NM_144670
|
A2ML1
|
alpha-2-macroglobulin-like 1
|
|
chr5_-_27074407
|
0.267
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr4_-_145159889
|
0.267
|
NM_002100
|
GYPB
|
glycophorin B (MNS blood group)
|
|
chr19_-_21438525
|
0.266
|
|
|
|
|
chr1_+_195503956
|
0.265
|
NM_001193640
NM_201253
|
CRB1
|
crumbs homolog 1 (Drosophila)
|
|
chr2_-_199922047
|
0.261
|
|
SATB2
|
SATB homeobox 2
|
|
chr11_+_89032112
|
0.261
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr10_+_118295417
|
0.261
|
NM_000936
|
PNLIP
|
pancreatic lipase
|
|
chr3_+_187840842
|
0.260
|
NM_014375
|
FETUB
|
fetuin B
|
|
chr5_+_140160966
|
0.259
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr14_-_60818263
|
0.259
|
NM_001017970
|
TMEM30B
|
transmembrane protein 30B
|
|
chr10_-_52315436
|
0.259
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr18_+_30589937
|
0.259
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr15_-_39151468
|
0.259
|
|
INO80
|
INO80 homolog (S. cerevisiae)
|
|
chr5_+_140719773
|
0.259
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr12_+_49522967
|
0.258
|
NM_182559
|
TMPRSS12
|
transmembrane (C-terminal) protease, serine 12
|
|
chr12_-_43593977
|
0.256
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr1_-_188713324
|
0.255
|
NM_199051
|
FAM5C
|
family with sequence similarity 5, member C
|
|
chr2_-_100088691
|
0.255
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr6_-_50124322
|
0.253
|
NM_001037498
|
DEFB112
|
defensin, beta 112
|
|
chr1_-_89414310
|
0.251
|
NM_207398
|
GBP7
|
guanylate binding protein 7
|
|
chr4_-_100575313
|
0.251
|
NM_001166504
NM_000673
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr10_-_69125932
|
0.249
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr2_+_190723216
|
0.249
|
NM_001042520
|
C2orf88
|
chromosome 2 open reading frame 88
|
|
chr12_+_77963563
|
0.248
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr4_+_155707089
|
0.247
|
|
FGB
|
fibrinogen beta chain
|
|
chr5_-_95774444
|
0.247
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|