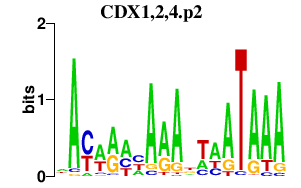
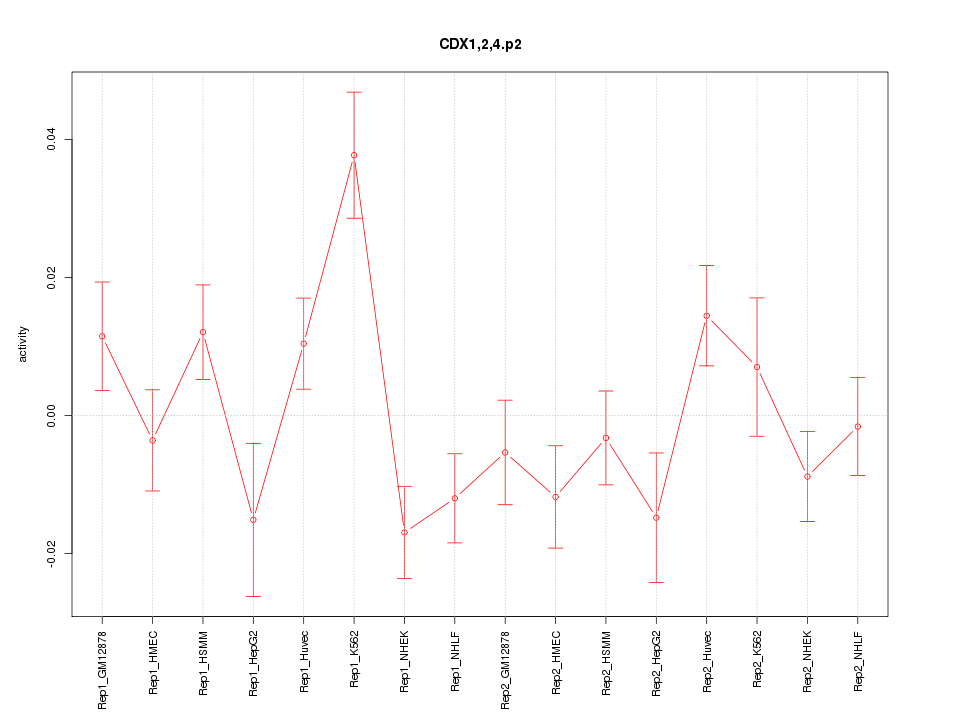
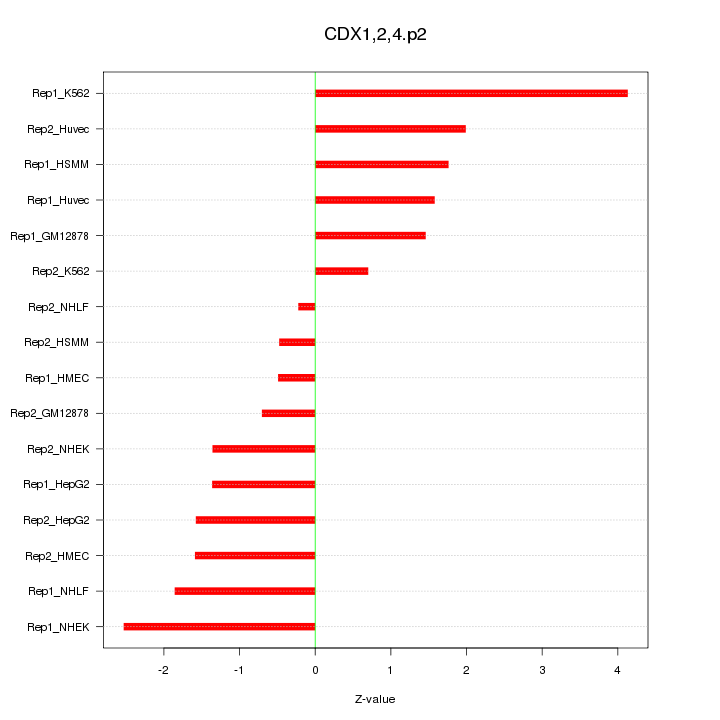
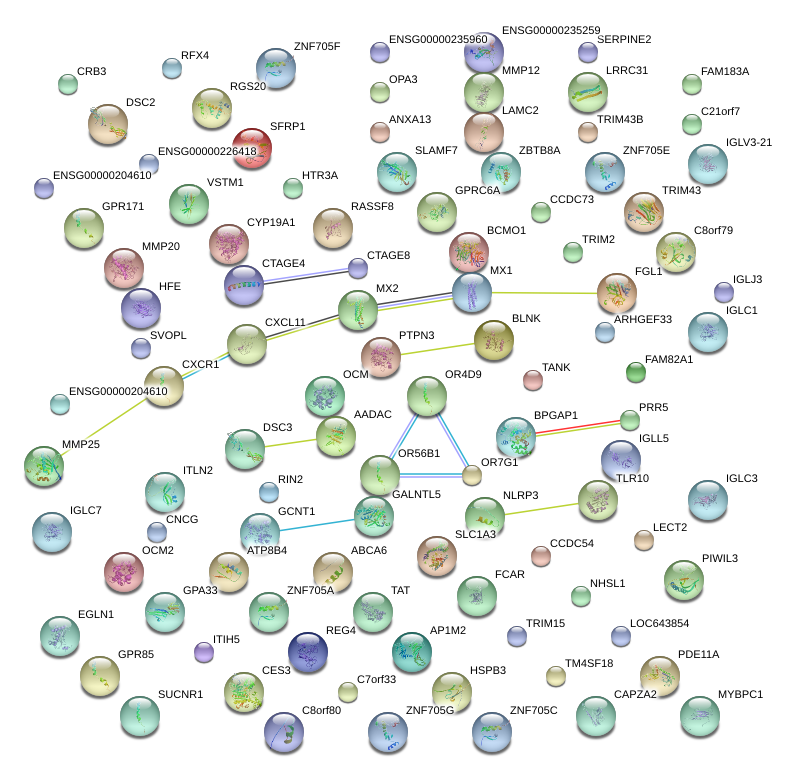
|
chr4_-_47678187
|
4.624
|
NM_001142564
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr7_+_151284396
|
4.015
|
NM_145292
|
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5
|
|
chr3_-_150533948
|
3.241
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr22_+_21384862
|
3.046
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr7_+_116238309
|
2.325
|
|
CAPZA2
|
capping protein (actin filament) muscle Z-line, alpha 2
|
|
chr8_-_7207899
|
2.296
|
NM_001164457
|
ZNF705G
|
zinc finger protein 705G
|
|
chr4_+_154397975
|
2.235
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr22_-_23500682
|
2.083
|
NM_001008496
|
PIWIL3
|
piwi-like 3 (Drosophila)
|
|
chr20_+_19818209
|
1.975
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr3_-_152403636
|
1.954
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr3_+_108578877
|
1.930
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr19_-_10558906
|
1.927
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr6_+_30238961
|
1.924
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr16_-_70168449
|
1.923
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr8_-_27997306
|
1.830
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr4_-_38460983
|
1.809
|
NM_001017388
NM_001195106
NM_001195107
NM_001195108
NM_030956
|
TLR10
|
toll-like receptor 10
|
|
chr22_+_43527101
|
1.806
|
NM_001017526
NM_001198726
NM_181335
|
ARHGAP8
|
Rho GTPase activating protein 8
|
|
chr1_+_32777358
|
1.790
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr16_+_65558878
|
1.694
|
NM_001185176
|
CES3
|
carboxylesterase 3
|
|
chr6_+_29963328
|
1.627
|
|
HLA-H
|
major histocompatibility complex, class I, H (pseudogene)
|
|
chr6_-_117256890
|
1.613
|
NM_148963
|
GPRC6A
|
G protein-coupled receptor, family C, group 6, member A
|
|
chr16_+_79829794
|
1.592
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr5_+_36644187
|
1.567
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr2_+_161701711
|
1.498
|
NM_004180
NM_133484
|
TANK
|
TRAF family member-associated NFKB activator
|
|
chr1_+_245647973
|
1.474
|
NM_001127462
NM_004895
NM_183395
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr5_-_156705250
|
1.420
|
NM_001001343
|
FNDC9
|
fibronectin type III domain containing 9
|
|
chr7_-_112513379
|
1.415
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr1_+_43386180
|
1.413
|
NM_001101376
|
FAM183A
|
family with sequence similarity 183, member A
|
|
chr11_-_102250876
|
1.403
|
NM_002426
|
MMP12
|
matrix metallopeptidase 12 (macrophage elastase)
|
|
chr6_-_25679197
|
1.378
|
|
|
|
|
chr10_-_98021144
|
1.351
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr19_-_59258846
|
1.346
|
NM_198481
|
VSTM1
|
V-set and transmembrane domain containing 1
|
|
chr2_+_38009405
|
1.321
|
NM_001170792
|
FAM82A1
|
family with sequence similarity 82, member A1
|
|
chr2_-_224574860
|
1.295
|
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr7_+_147918589
|
1.285
|
NM_145304
|
C7orf33
|
chromosome 7 open reading frame 33
|
|
chr2_-_218739891
|
1.269
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr4_-_77176163
|
1.266
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr18_-_26935883
|
1.216
|
NM_004949
NM_024422
|
DSC2
|
desmocollin 2
|
|
chr11_+_59038961
|
1.209
|
NM_001004711
|
OR4D9
|
olfactory receptor, family 4, subfamily D, member 9
|
|
chr21_+_41719847
|
1.193
|
NM_002462
NM_001178046
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse)
|
|
chr11_+_113351006
|
1.160
|
NM_000869
NM_213621
|
HTR3A
|
5-hydroxytryptamine (serotonin) receptor 3A
|
|
chr10_-_7699192
|
1.151
|
|
ITIH5
|
inter-alpha (globulin) inhibitor H5
|
|
chr11_-_32772731
|
1.150
|
NM_001008391
|
CCDC73
|
coiled-coil domain containing 73
|
|
chr8_-_124818754
|
1.142
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr2_-_178495768
|
1.141
|
NM_001077358
|
PDE11A
|
phosphodiesterase 11A
|
|
chr7_+_142978994
|
1.132
|
NM_001008747
|
CTAGE6P
CTAGE15P
|
CTAGE family, member 6, pseudogene
CTAGE family, member 15, pseudogene
|
|
chr5_-_135318420
|
1.098
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr1_-_229627412
|
1.093
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr1_-_178100204
|
1.092
|
|
|
|
|
chr19_+_6415259
|
1.088
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr3_-_171070314
|
1.084
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr2_+_95621492
|
1.078
|
NM_138800
|
TRIM43
|
tripartite motif containing 43
|
|
chr12_+_26096757
|
1.075
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_-_142536082
|
1.073
|
|
|
|
|
chr1_+_181421796
|
1.042
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr12_+_100512877
|
1.042
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr1_+_158975662
|
0.995
|
NM_021181
|
SLAMF7
|
SLAM family member 7
|
|
chr21_+_29439768
|
0.992
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr3_+_153074118
|
0.983
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr1_-_159191184
|
0.982
|
NM_080878
|
ITLN2
|
intelectin 2
|
|
chr7_-_97457286
|
0.978
|
NM_006188
|
OCM2
|
oncomodulin 2
|
|
chr1_-_120155581
|
0.969
|
NM_001159352
NM_001159353
NM_032044
|
REG4
|
regenerating islet-derived family, member 4
|
|
chr8_-_17787342
|
0.955
|
|
FGL1
|
fibrinogen-like 1
|
|
chr19_+_60077360
|
0.950
|
NM_002000
NM_133269
NM_133271
NM_133272
NM_133273
NM_133274
NM_133277
NM_133278
NM_133279
|
FCAR
|
Fc fragment of IgA, receptor for
|
|
chr15_-_49418027
|
0.944
|
NM_000103
NM_031226
|
CYP19A1
|
cytochrome P450, family 19, subfamily A, polypeptide 1
|
|
chr3_+_153014550
|
0.939
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr7_-_138014329
|
0.939
|
NM_001139456
|
SVOPL
|
SVOP-like
|
|
chr15_-_48192804
|
0.936
|
|
ATP8B4
|
ATPase, class I, type 8B, member 4
|
|
chr12_+_105602606
|
0.924
|
NM_032491
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr1_-_165326148
|
0.922
|
NM_005814
|
GPA33
|
glycoprotein A33 (transmembrane)
|
|
chr11_+_5714253
|
0.913
|
NM_001005180
|
OR56B1
|
olfactory receptor, family 56, subfamily B, member 1
|
|
chr8_+_12914143
|
0.906
|
NM_001099677
|
KIAA1456
|
KIAA1456
|
|
chr9_-_111253484
|
0.901
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr5_+_53787187
|
0.897
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr3_+_46628928
|
0.894
|
NM_001195442
|
LOC100132146
|
hypothetical LOC100132146
|
|
chr11_-_102001259
|
0.892
|
NM_004771
|
MMP20
|
matrix metallopeptidase 20
|
|
chr2_+_39000007
|
0.891
|
NM_001145451
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33
|
|
chr6_-_138935359
|
0.876
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr19_-_9087438
|
0.870
|
NM_001005192
|
OR7G1
|
olfactory receptor, family 7, subfamily G, member 1
|
|
chr9_+_78305368
|
0.848
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr1_-_143232526
|
0.848
|
|
FAM91A2
|
family with sequence similarity 91, member A2
|
|
chr8_-_41286088
|
0.840
|
NM_003012
|
SFRP1
|
secreted frizzled-related protein 1
|
|
chr4_-_77163609
|
0.833
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr1_-_146543102
|
0.832
|
|
|
|
|
chr1_+_242283129
|
0.830
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_218768170
|
0.828
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr1_-_181889070
|
0.820
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr6_+_158990982
|
0.820
|
NM_001009991
|
SYTL3
|
synaptotagmin-like 3
|
|
chr19_-_5797465
|
0.809
|
|
FUT3
|
fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group)
|
|
chr12_-_51065629
|
0.804
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr10_-_23673777
|
0.798
|
NM_153714
|
C10orf67
|
chromosome 10 open reading frame 67
|
|
chr5_-_172016174
|
0.796
|
|
|
|
|
chr19_-_49725387
|
0.796
|
NM_001102597
NM_001102598
NM_001102599
NM_001102600
|
CEACAM20
|
carcinoembryonic antigen-related cell adhesion molecule 20
|
|
chr6_+_150304813
|
0.795
|
NM_025217
|
ULBP2
|
UL16 binding protein 2
|
|
chr19_+_55488632
|
0.783
|
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr15_+_57696200
|
0.780
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr11_-_102081654
|
0.780
|
NM_022122
|
MMP27
|
matrix metallopeptidase 27
|
|
chr12_-_10983072
|
0.779
|
NM_023922
|
TAS2R14
|
taste receptor, type 2, member 14
|
|
chr12_+_53901075
|
0.777
|
NM_001005280
|
OR10A7
|
olfactory receptor, family 10, subfamily A, member 7
|
|
chr22_-_20667131
|
0.776
|
NM_003935
|
TOP3B
|
topoisomerase (DNA) III beta
|
|
chr10_-_62002672
|
0.772
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr19_+_40895669
|
0.769
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr4_+_71418886
|
0.769
|
NM_212557
|
AMTN
|
amelotin
|
|
chr18_+_606671
|
0.769
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr8_-_17797127
|
0.767
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr1_-_23393808
|
0.767
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr1_+_207944807
|
0.761
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr9_+_102380156
|
0.758
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr10_-_62163083
|
0.757
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr11_+_110890719
|
0.755
|
NM_001100388
NM_207430
|
C11orf88
|
chromosome 11 open reading frame 88
|
|
chr13_+_77007809
|
0.755
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr7_+_134114912
|
0.745
|
|
CALD1
|
caldesmon 1
|
|
chr3_-_17716515
|
0.741
|
NM_001134381
|
TBC1D5
|
TBC1 domain family, member 5
|
|
chr20_+_57966865
|
0.741
|
NM_177980
|
CDH26
|
cadherin 26
|
|
chr1_+_61697209
|
0.737
|
|
NFIA
|
nuclear factor I/A
|
|
chr13_+_97856148
|
0.736
|
|
|
|
|
chr4_+_41678469
|
0.732
|
NM_001029955
|
DCAF4L1
|
DDB1 and CUL4 associated factor 4-like 1
|
|
chr22_-_20667165
|
0.731
|
|
TOP3B
|
topoisomerase (DNA) III beta
|
|
chr7_+_134114957
|
0.727
|
|
CALD1
|
caldesmon 1
|
|
chr10_-_48010879
|
0.725
|
NM_002900
|
RBP3
|
retinol binding protein 3, interstitial
|
|
chr12_-_121753795
|
0.721
|
NM_177551
|
GPR109A
|
G protein-coupled receptor 109A
|
|
chr22_-_20667211
|
0.721
|
|
TOP3B
|
topoisomerase (DNA) III beta
|
|
chr20_-_49747397
|
0.719
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr17_-_36409632
|
0.718
|
NM_031959
|
KRTAP3-2
|
keratin associated protein 3-2
|
|
chr2_+_98070026
|
0.718
|
NM_144992
|
VWA3B
|
von Willebrand factor A domain containing 3B
|
|
chr1_-_67366807
|
0.716
|
NM_001013674
|
C1orf141
|
chromosome 1 open reading frame 141
|
|
chr11_+_60016826
|
0.715
|
NM_001164470
NM_017716
|
MS4A12
|
membrane-spanning 4-domains, subfamily A, member 12
|
|
chr1_+_195123766
|
0.715
|
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr22_-_43987978
|
0.714
|
|
KIAA0930
|
KIAA0930
|
|
chr6_-_130585650
|
0.713
|
NM_001017373
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr5_+_127012611
|
0.707
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr12_-_51132129
|
0.705
|
NM_005555
|
KRT6B
|
keratin 6B
|
|
chr6_-_29451042
|
0.703
|
NM_030959
|
OR12D3
|
olfactory receptor, family 12, subfamily D, member 3
|
|
chr17_+_3326045
|
0.694
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr11_-_64484184
|
0.694
|
NM_001037225
|
C11orf85
|
chromosome 11 open reading frame 85
|
|
chr5_+_140772691
|
0.693
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr4_+_37638450
|
0.693
|
NM_006607
|
PTTG2
|
pituitary tumor-transforming 2
|
|
chr1_+_43837096
|
0.688
|
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr16_+_67235651
|
0.687
|
NM_001793
|
CDH3
|
cadherin 3, type 1, P-cadherin (placental)
|
|
chr11_+_73339011
|
0.687
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr6_+_29663661
|
0.679
|
NM_007160
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2
|
|
chr3_+_133801670
|
0.674
|
NM_178445
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr10_-_27097782
|
0.673
|
|
ABI1
|
abl-interactor 1
|
|
chr10_-_30958631
|
0.672
|
NM_183058
|
LYZL2
|
lysozyme-like 2
|
|
chr12_+_8557402
|
0.672
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr9_+_140164359
|
0.671
|
|
TUBBP5
|
tubulin, beta pseudogene 5
|
|
chr5_-_180099663
|
0.667
|
NM_001001657
|
OR2Y1
|
olfactory receptor, family 2, subfamily Y, member 1
|
|
chr2_+_201182187
|
0.655
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr12_+_8216416
|
0.643
|
NM_001004328
|
ZNF705A
|
zinc finger protein 705A
|
|
chr11_-_16953135
|
0.640
|
|
RPL36A
|
ribosomal protein L36a
|
|
chr10_+_74323344
|
0.640
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr7_-_38279757
|
0.627
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr1_+_218768146
|
0.627
|
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr7_-_111216180
|
0.623
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr2_+_88605283
|
0.623
|
NM_152670
|
C2orf51
|
chromosome 2 open reading frame 51
|
|
chr5_+_68823874
|
0.620
|
NM_002538
|
OCLN
|
occludin
|
|
chr3_+_12367993
|
0.619
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr7_+_101810018
|
0.618
|
|
|
|
|
chrX_-_62487693
|
0.616
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chr12_-_25692754
|
0.615
|
NM_001145727
|
IFLTD1
|
intermediate filament tail domain containing 1
|
|
chr1_-_145163779
|
0.612
|
NM_001144829
NM_001144830
NM_001461
|
FMO5
|
flavin containing monooxygenase 5
|
|
chr8_+_92330691
|
0.610
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr12_+_10222823
|
0.609
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr3_+_38282301
|
0.609
|
NM_004256
|
SLC22A13
|
solute carrier family 22 (organic anion transporter), member 13
|
|
chr7_+_142629251
|
0.609
|
NM_176882
|
TAS2R40
|
taste receptor, type 2, member 40
|
|
chr4_+_89519004
|
0.602
|
|
HERC6
|
hect domain and RLD 6
|
|
chr6_-_38671741
|
0.598
|
NM_001172418
NM_152733
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr1_-_54256390
|
0.597
|
NM_001010978
|
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1
|
|
chr12_-_27815475
|
0.592
|
NM_001146221
|
LOC100287284
|
MANSC domain-containing protein ENSP00000370673
|
|
chr15_-_70385709
|
0.590
|
NM_001172685
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr11_+_113165134
|
0.590
|
|
LOC390251
|
SH3-domain GRB2-like 1 pseudogene
|
|
chr1_-_177106702
|
0.588
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr15_-_68781662
|
0.587
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr6_+_151688327
|
0.585
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr4_+_89518898
|
0.581
|
NM_001165136
NM_017912
|
HERC6
|
hect domain and RLD 6
|
|
chr15_+_19869841
|
0.580
|
NM_001004719
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2
|
|
chr10_+_71482542
|
0.579
|
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr2_+_113532685
|
0.573
|
NM_173170
|
IL1F5
|
interleukin 1 family, member 5 (delta)
|
|
chr12_-_121767272
|
0.569
|
NM_006018
|
GPR109B
|
G protein-coupled receptor 109B
|
|
chr2_-_26395420
|
0.566
|
NM_001145168
NM_001145169
|
GPR113
|
G protein-coupled receptor 113
|
|
chr4_+_155707089
|
0.565
|
|
FGB
|
fibrinogen beta chain
|
|
chr7_+_137411717
|
0.564
|
NM_001190906
NM_001190907
NM_005989
|
AKR1D1
|
aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase)
|
|
chr20_-_33783409
|
0.559
|
|
RBM39
|
RNA binding motif protein 39
|
|
chr6_+_29732736
|
0.559
|
NM_001008228
NM_001008229
NM_001170418
NM_002433
NM_206809
NM_206810
NM_206811
NM_206812
NM_206814
|
MOG
|
myelin oligodendrocyte glycoprotein
|
|
chr3_-_140879463
|
0.555
|
NM_178177
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr4_-_5881271
|
0.547
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr3_-_152449591
|
0.546
|
NM_001081455
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chr12_+_55443358
|
0.545
|
NM_003725
|
HSD17B6
|
hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse)
|
|
chr12_-_11106159
|
0.544
|
NM_176887
|
TAS2R46
|
taste receptor, type 2, member 46
|
|
chr6_-_136889302
|
0.540
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr1_+_195503956
|
0.536
|
NM_001193640
NM_201253
|
CRB1
|
crumbs homolog 1 (Drosophila)
|
|
chr7_+_75928907
|
0.535
|
NM_001102594
NM_001102595
NM_020892
|
DTX2
|
deltex homolog 2 (Drosophila)
|
|
chr9_-_103397023
|
0.532
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr19_-_56946911
|
0.531
|
NM_001193306
NM_002029
|
FPR1
|
formyl peptide receptor 1
|
|
chr3_+_46591048
|
0.531
|
NM_001174136
|
TDGF1
|
teratocarcinoma-derived growth factor 1
|
|
chr11_-_129268113
|
0.530
|
NM_006165
|
NFRKB
|
nuclear factor related to kappaB binding protein
|
|
chr11_-_111636792
|
0.525
|
NM_001145024
|
C11orf34
|
chromosome 11 open reading frame 34
|
|
chr19_+_50144267
|
0.520
|
|
APOC2
|
apolipoprotein C-II
|
|
chr9_+_124463665
|
0.519
|
NM_001005236
|
OR1L1
|
olfactory receptor, family 1, subfamily L, member 1
|
|
chr12_+_74014685
|
0.517
|
NM_152779
|
GLIPR1L1
|
GLI pathogenesis-related 1 like 1
|