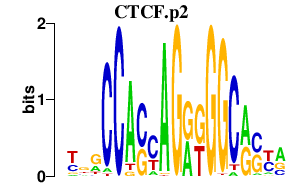
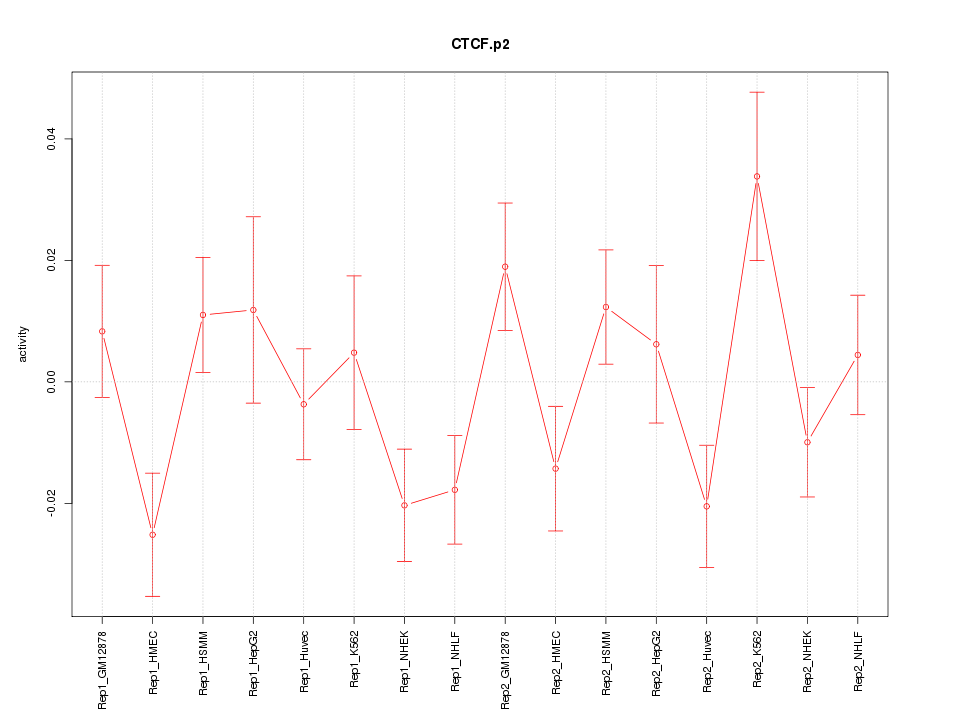
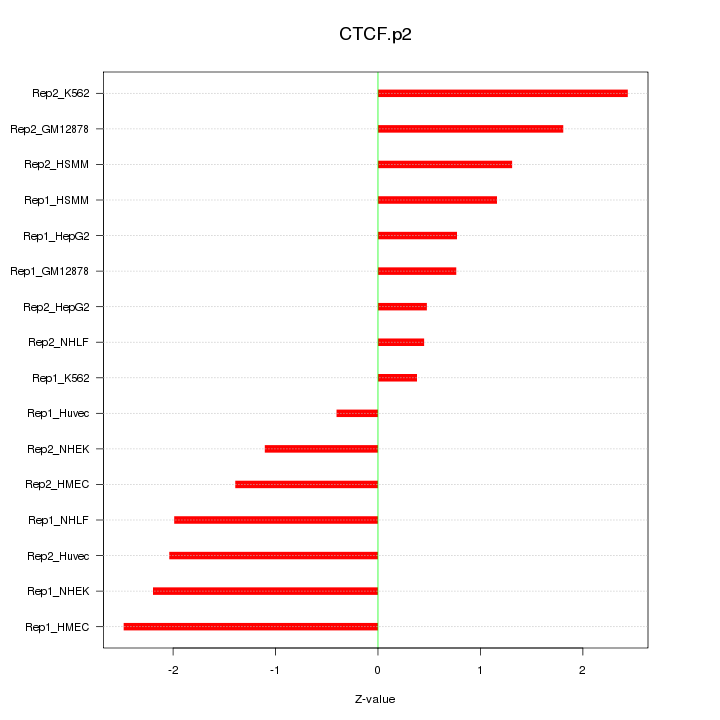
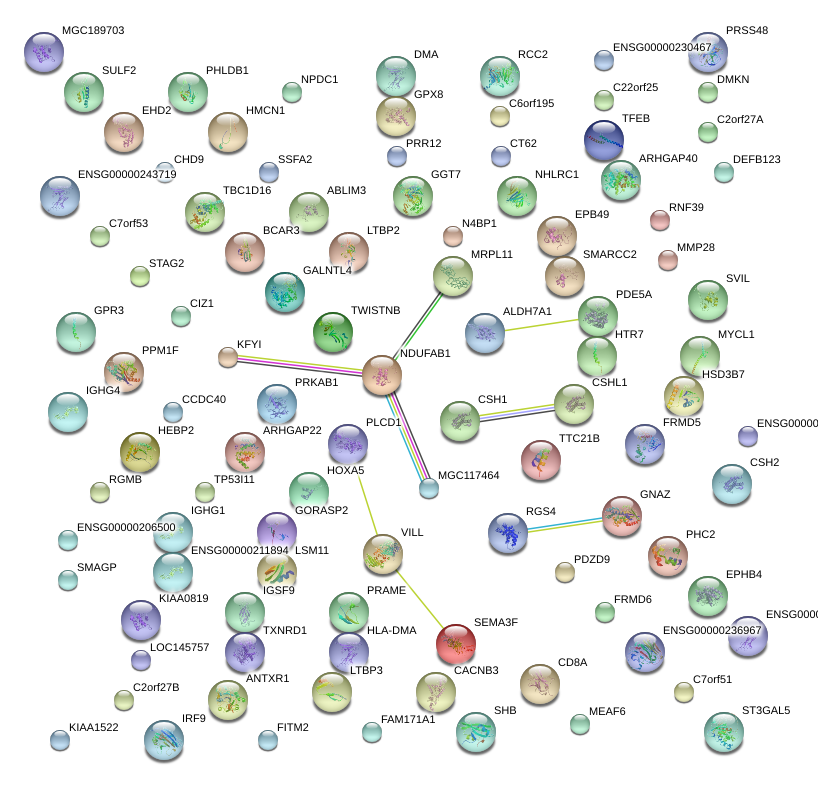
|
chr6_-_2580296
|
1.727
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr20_+_36663865
|
1.484
|
NM_001164431
|
ARHGAP40
|
Rho GTPase activating protein 40
|
|
chr19_-_40693176
|
1.451
|
NM_001126059
|
DMKN
|
dermokine
|
|
chr22_-_16887151
|
1.439
|
NM_001122731
NM_001136004
NM_015241
|
MICAL3
|
microtubule associated monoxygenase, calponin and LIM domain containing 3
|
|
chr5_+_157103280
|
1.358
|
NM_173491
|
LSM11
|
LSM11, U7 small nuclear RNA associated
|
|
chr2_+_69093641
|
1.341
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr1_+_183970093
|
1.280
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr17_-_31146660
|
1.239
|
NM_001032278
NM_024302
|
MMP28
|
matrix metallopeptidase 28
|
|
chr1_-_158182006
|
1.196
|
NM_001135050
NM_020789
|
IGSF9
|
immunoglobulin superfamily, member 9
|
|
chr1_-_33669190
|
1.190
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr5_-_125958503
|
1.176
|
NM_001182
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr14_+_51188289
|
1.161
|
NM_152330
|
FRMD6
|
FERM domain containing 6
|
|
chr15_-_69194854
|
1.143
|
NM_001102658
|
CT62
|
cancer/testis antigen 62
|
|
chr22_+_21742364
|
1.128
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr15_-_42274529
|
1.097
|
NM_032892
|
FRMD5
|
FERM domain containing 5
|
|
chr10_-_49534304
|
1.095
|
|
ARHGAP22
|
Rho GTPase activating protein 22
|
|
chr5_+_148501178
|
1.093
|
NM_014945
|
ABLIM3
|
actin binding LIM protein family, member 3
|
|
chr22_+_21742727
|
1.018
|
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr2_-_86889029
|
1.005
|
NM_001145873
|
CD8A
|
CD8a molecule
|
|
chr6_+_139055244
|
0.945
|
|
FLJ46906
|
hypothetical LOC441172
|
|
chr20_-_45848684
|
0.941
|
NM_001161841
NM_198596
|
SULF2
|
sulfatase 2
|
|
chr3_+_50167424
|
0.916
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr22_-_21231421
|
0.908
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr1_-_40140126
|
0.895
|
NM_001033081
NM_001033082
NM_005376
|
MYCL1
|
v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian)
|
|
chr6_-_18230829
|
0.890
|
NM_198586
|
NHLRC1
|
NHL repeat containing 1
|
|
chr16_-_21919928
|
0.885
|
NM_173806
|
PDZD9
|
PDZ domain containing 9
|
|
chr20_-_42373151
|
0.881
|
NM_001080472
|
FITM2
|
fat storage-inducing transmembrane protein 2
|
|
chr17_-_75624048
|
0.860
|
NM_019020
|
TBC1D16
|
TBC1 domain family, member 16
|
|
chr10_-_92607397
|
0.859
|
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled)
|
|
chr15_+_99207555
|
0.842
|
|
LOC145757
|
hypothetical LOC145757
|
|
chr10_-_30064600
|
0.827
|
NM_003174
|
SVIL
|
supervillin
|
|
chr6_+_6670117
|
0.824
|
|
|
|
|
chr5_+_98132898
|
0.817
|
NM_001012761
|
RGMB
|
RGM domain family, member B
|
|
chr14_-_74148422
|
0.807
|
NM_000428
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr12_-_49950236
|
0.804
|
NM_001033873
NM_001031628
|
SMAGP
|
small cell adhesion glycoprotein
|
|
chr15_-_42274760
|
0.800
|
|
FRMD5
|
FERM domain containing 5
|
|
chr12_+_118590039
|
0.799
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr20_+_29492071
|
0.795
|
NM_153324
|
DEFB123
|
defensin, beta 123
|
|
chr16_-_23515104
|
0.793
|
NM_005003
|
NDUFAB1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa
|
|
chr17_-_59342319
|
0.785
|
NM_001318
NM_022579
NM_022580
NM_022581
|
CSHL1
|
chorionic somatomammotropin hormone-like 1
|
|
chr5_+_54491702
|
0.782
|
NM_001008397
|
GPX8
|
glutathione peroxidase 8 (putative)
|
|
chr1_+_32979930
|
0.763
|
NM_020888
|
KIAA1522
|
KIAA1522
|
|
chrX_+_122922058
|
0.750
|
NM_001042749
|
STAG2
|
stromal antigen 2
|
|
chr6_-_30151526
|
0.740
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr2_+_171493870
|
0.739
|
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr16_+_30903963
|
0.738
|
NM_001142777
NM_001142778
NM_025193
|
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr12_+_103133631
|
0.732
|
NM_001093771
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr17_+_75625025
|
0.732
|
NM_017950
|
CCDC40
|
coiled-coil domain containing 40
|
|
chr2_-_85969527
|
0.720
|
NM_003896
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5
|
|
chr22_+_21027538
|
0.717
|
|
|
|
|
chr19_+_52908412
|
0.714
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr9_-_38059207
|
0.702
|
NM_003028
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr2_+_171494026
|
0.700
|
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr12_+_47494882
|
0.685
|
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr12_+_62523648
|
0.675
|
|
|
|
|
chr14_-_106154332
|
0.671
|
|
LOC100126583
IGHG1
|
hypothetical LOC100126583
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr4_+_152417774
|
0.663
|
NM_183375
|
PRSS48
|
protease, serine, 48
|
|
chr22_-_20637188
|
0.656
|
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F
|
|
chr6_-_33028790
|
0.655
|
NM_006120
|
HLA-DMA
|
major histocompatibility complex, class II, DM alpha
|
|
chr1_-_17638797
|
0.652
|
NM_018715
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr11_-_44928803
|
0.651
|
NM_001076787
NM_006034
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr7_-_27149750
|
0.636
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr16_+_51646292
|
0.634
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr1_+_161308318
|
0.632
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr11_-_65962836
|
0.628
|
NM_016050
NM_170738
NM_170739
|
MRPL11
|
mitochondrial ribosomal protein L11
|
|
chr2_+_182464910
|
0.627
|
|
SSFA2
|
sperm specific antigen 2
|
|
chr2_-_166518500
|
0.613
|
NM_024753
|
TTC21B
|
tetratricopeptide repeat domain 21B
|
|
chr9_-_129993615
|
0.612
|
NM_001131016
NM_001131017
NM_001131018
|
CIZ1
|
CDKN1A interacting zinc finger protein 1
|
|
chr1_+_27591738
|
0.607
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr1_-_37752925
|
0.599
|
NM_022756
|
MEAF6
|
MYST/Esa1-associated factor 6
|
|
chr6_+_138767151
|
0.587
|
|
HEBP2
|
heme binding protein 2
|
|
chr3_+_38010618
|
0.584
|
|
VILL
|
villin-like
|
|
chr10_-_92607650
|
0.575
|
NM_000872
NM_019859
NM_019860
|
HTR7
|
5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled)
|
|
chr20_-_32924219
|
0.570
|
NM_178026
|
GGT7
|
gamma-glutamyltransferase 7
|
|
chr12_-_54869511
|
0.563
|
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr9_-_139060416
|
0.563
|
NM_015392
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr1_-_93919789
|
0.561
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr3_-_38046034
|
0.561
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr9_-_38059082
|
0.560
|
|
SHB
|
Src homology 2 domain containing adaptor protein B
|
|
chr4_-_140436401
|
0.546
|
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr7_+_99919485
|
0.543
|
NM_173564
|
C7orf51
|
chromosome 7 open reading frame 51
|
|
chr7_-_19715127
|
0.542
|
NM_001002926
|
TWISTNB
|
TWIST neighbor
|
|
chr19_+_54786723
|
0.540
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr11_+_117983515
|
0.530
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr15_-_37999519
|
0.526
|
|
|
|
|
chr7_+_111908143
|
0.524
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr7_-_100262971
|
0.521
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr22_+_18388633
|
0.518
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr2_-_132275703
|
0.517
|
NM_214461
|
C2orf27B
|
chromosome 2 open reading frame 27B
|
|
chr6_-_41781655
|
0.516
|
|
TFEB
|
transcription factor EB
|
|
chr11_+_111666133
|
0.513
|
|
|
|
|
chr10_-_15453060
|
0.509
|
NM_001010924
|
FAM171A1
|
family with sequence similarity 171, member A1
|
|
chr16_-_47201588
|
0.508
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr14_+_23700261
|
0.506
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr8_+_21980315
|
0.506
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr11_-_11600118
|
0.505
|
NM_198516
|
GALNTL4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chr4_-_120767777
|
0.503
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr6_+_35544155
|
0.503
|
NM_007104
|
RPL10A
|
ribosomal protein L10a
|
|
chr9_+_140164359
|
0.501
|
|
TUBBP5
|
tubulin, beta pseudogene 5
|
|
chr9_-_111123017
|
0.492
|
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chr7_-_122313646
|
0.485
|
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr12_+_106236281
|
0.485
|
NM_001018072
|
BTBD11
|
BTB (POZ) domain containing 11
|
|
chr3_+_49183983
|
0.484
|
NM_173546
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr5_-_146813343
|
0.482
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr7_-_122314017
|
0.473
|
NM_001009571
NM_001167940
NM_017954
|
CADPS2
|
Ca++-dependent secretion activator 2
|
|
chr22_+_21393107
|
0.470
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr11_-_128281309
|
0.467
|
|
C11orf45
|
chromosome 11 open reading frame 45
|
|
chr17_+_21249040
|
0.467
|
NM_001194958
|
KCNJ18
|
potassium inwardly-rectifying channel, subfamily J, member 18
|
|
chr22_-_20637212
|
0.466
|
NM_014634
|
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F
|
|
chr10_+_102749222
|
0.466
|
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr6_+_35544174
|
0.466
|
|
RPL10A
|
ribosomal protein L10a
|
|
chr1_+_64012154
|
0.466
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr6_-_58395610
|
0.466
|
|
GUSBP4
|
glucuronidase, beta pseudogene 4
|
|
chr6_-_41855564
|
0.465
|
NM_006653
|
FRS3
|
fibroblast growth factor receptor substrate 3
|
|
chr17_-_24253821
|
0.463
|
NM_144683
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13
|
|
chr19_-_18913017
|
0.462
|
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr13_-_39075304
|
0.457
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr19_-_18913032
|
0.456
|
NM_004838
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr19_-_18913011
|
0.453
|
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr11_-_64906683
|
0.453
|
NM_001077241
NM_182556
|
SLC25A45
|
solute carrier family 25, member 45
|
|
chr6_+_25387273
|
0.449
|
|
LRRC16A
|
leucine rich repeat containing 16A
|
|
chr7_-_91347915
|
0.443
|
NM_006980
|
MTERF
|
mitochondrial transcription termination factor
|
|
chr1_+_46418786
|
0.442
|
|
TSPAN1
|
tetraspanin 1
|
|
chr8_-_91066060
|
0.440
|
NM_002485
|
NBN
|
nibrin
|
|
chr20_+_28225511
|
0.440
|
|
LOC283788
FRG1B
|
FSHD region gene 1 pseudogene
FSHD region gene 1 family, member B
|
|
chr12_+_50104859
|
0.434
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr22_-_29113301
|
0.433
|
NM_001017981
|
RNF215
|
ring finger protein 215
|
|
chr17_-_19290745
|
0.433
|
|
|
|
|
chr11_+_60448487
|
0.433
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr12_+_46643592
|
0.432
|
NM_001143841
NM_001143842
NM_001143843
NM_024056
|
TMEM106C
|
transmembrane protein 106C
|
|
chr13_+_30672087
|
0.432
|
NM_194318
|
B3GALTL
|
beta 1,3-galactosyltransferase-like
|
|
chr2_-_20714303
|
0.431
|
NM_022460
|
HS1BP3
|
HCLS1 binding protein 3
|
|
chr2_-_175060001
|
0.431
|
NM_001033045
NM_152529
|
GPR155
|
G protein-coupled receptor 155
|
|
chr12_-_54869568
|
0.431
|
NM_001130420
NM_003075
NM_139067
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr6_+_127629677
|
0.425
|
NM_030963
|
RNF146
|
ring finger protein 146
|
|
chr16_-_11798507
|
0.422
|
|
ZC3H7A
|
zinc finger CCCH-type containing 7A
|
|
chr2_-_39310176
|
0.420
|
NM_001009565
|
CDKL4
|
cyclin-dependent kinase-like 4
|
|
chr6_+_25387630
|
0.420
|
NM_001173977
NM_017640
|
LRRC16A
|
leucine rich repeat containing 16A
|
|
chr20_+_31783426
|
0.416
|
NM_032819
|
ZNF341
|
zinc finger protein 341
|
|
chr10_-_101759630
|
0.416
|
NM_015221
|
DNMBP
|
dynamin binding protein
|
|
chr5_-_70621688
|
0.415
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr1_-_6537139
|
0.414
|
|
NOL9
|
nucleolar protein 9
|
|
chr7_+_28305464
|
0.413
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_-_49949999
|
0.412
|
|
SMAGP
|
small cell adhesion glycoprotein
|
|
chr7_+_75669130
|
0.410
|
NM_001110199
|
SRRM3
|
serine/arginine repetitive matrix 3
|
|
chr9_-_129537372
|
0.409
|
NM_001085347
NM_001134430
NM_001134431
NM_130459
|
TOR2A
|
torsin family 2, member A
|
|
chr9_-_132804243
|
0.409
|
NM_001145106
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr22_+_18388602
|
0.408
|
NM_152906
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr19_-_18912899
|
0.405
|
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr1_-_17638729
|
0.405
|
|
RCC2
|
regulator of chromosome condensation 2
|
|
chr1_-_6537171
|
0.401
|
NM_024654
|
NOL9
|
nucleolar protein 9
|
|
chr9_+_126655516
|
0.398
|
NM_001045476
|
WDR38
|
WD repeat domain 38
|
|
chr3_-_48573556
|
0.392
|
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr9_+_33614214
|
0.390
|
|
ANXA2P2
|
annexin A2 pseudogene 2
|
|
chr7_-_99517128
|
0.385
|
NM_017715
NM_032924
|
ZNF3
|
zinc finger protein 3
|
|
chr1_+_9217444
|
0.382
|
NM_004285
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr6_+_3794620
|
0.378
|
NM_012135
|
FAM50B
|
family with sequence similarity 50, member B
|
|
chr13_-_112308729
|
0.378
|
|
|
|
|
chr2_-_190892730
|
0.377
|
|
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase
|
|
chr4_-_24590867
|
0.376
|
NM_173463
|
CCDC149
|
coiled-coil domain containing 149
|
|
chr8_-_91065961
|
0.375
|
|
NBN
|
nibrin
|
|
chr10_+_6284845
|
0.367
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr3_+_115040369
|
0.366
|
NM_017577
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr17_-_19206475
|
0.365
|
NM_015681
|
B9D1
|
B9 protein domain 1
|
|
chr19_+_45168872
|
0.362
|
NM_006503
NM_153001
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4
|
|
chr14_+_104226769
|
0.359
|
NM_001031714
NM_022489
NM_032714
|
INF2
|
inverted formin, FH2 and WH2 domain containing
|
|
chr5_-_118352122
|
0.358
|
NM_173666
|
DTWD2
|
DTW domain containing 2
|
|
chr2_+_182464713
|
0.357
|
NM_001130445
NM_006751
|
SSFA2
|
sperm specific antigen 2
|
|
chr7_-_22506320
|
0.356
|
NM_001164460
NM_207342
|
MGC87042
|
STEAP family protein MGC87042
|
|
chr9_-_111122826
|
0.355
|
NM_018424
NM_019114
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chr17_-_58877276
|
0.353
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr17_-_69869552
|
0.353
|
NM_001080466
|
BTBD17
|
BTB (POZ) domain containing 17
|
|
chr20_-_1395463
|
0.352
|
|
NSFL1C
|
NSFL1 (p97) cofactor (p47)
|
|
chr11_+_107384617
|
0.349
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr17_-_52393364
|
0.345
|
NM_004645
|
COIL
|
coilin
|
|
chr12_+_13088412
|
0.344
|
|
KIAA1467
|
KIAA1467
|
|
chr6_-_47385470
|
0.340
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr19_+_47480445
|
0.339
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr14_-_106084085
|
0.339
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr6_-_47385605
|
0.336
|
NM_014452
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr11_+_118332217
|
0.335
|
NM_006760
|
UPK2
|
uroplakin 2
|
|
chr12_-_119187871
|
0.335
|
|
PXN
|
paxillin
|
|
chr2_-_218865451
|
0.335
|
NM_022152
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1
|
|
chr17_+_72376022
|
0.333
|
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr20_+_42537927
|
0.332
|
NM_001039199
NM_024331
|
TTPAL
|
tocopherol (alpha) transfer protein-like
|
|
chr8_+_43067817
|
0.329
|
|
|
|
|
chr17_-_43263859
|
0.322
|
NM_145255
NM_148887
|
MRPL10
|
mitochondrial ribosomal protein L10
|
|
chr8_+_43067805
|
0.321
|
NM_032237
|
SGK196
|
protein kinase-like protein SgK196
|
|
chr6_-_47385599
|
0.320
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr22_+_18388656
|
0.319
|
|
C22orf25
|
chromosome 22 open reading frame 25
|
|
chr20_+_48240867
|
0.318
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr1_+_9217384
|
0.318
|
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr11_+_66841885
|
0.318
|
|
LOC100130987
|
similar to hCG1815675
|
|
chr11_+_67130898
|
0.316
|
NM_001166102
NM_007103
|
NDUFV1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa
|
|
chrX_+_11686198
|
0.314
|
NM_078628
NM_078629
|
MSL3
|
male-specific lethal 3 homolog (Drosophila)
|
|
chr12_+_121803255
|
0.311
|
NM_003677
|
DENR
|
density-regulated protein
|
|
chr10_+_73241645
|
0.311
|
NM_001171935
NM_001171936
|
CDH23
|
cadherin-related 23
|
|
chr6_-_47385604
|
0.310
|
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21
|
|
chr9_+_696744
|
0.309
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr18_+_586997
|
0.309
|
NM_014410
|
CLUL1
|
clusterin-like 1 (retinal)
|