|
chr8_-_114518194
|
2.748
|
NM_052900
NM_198123
|
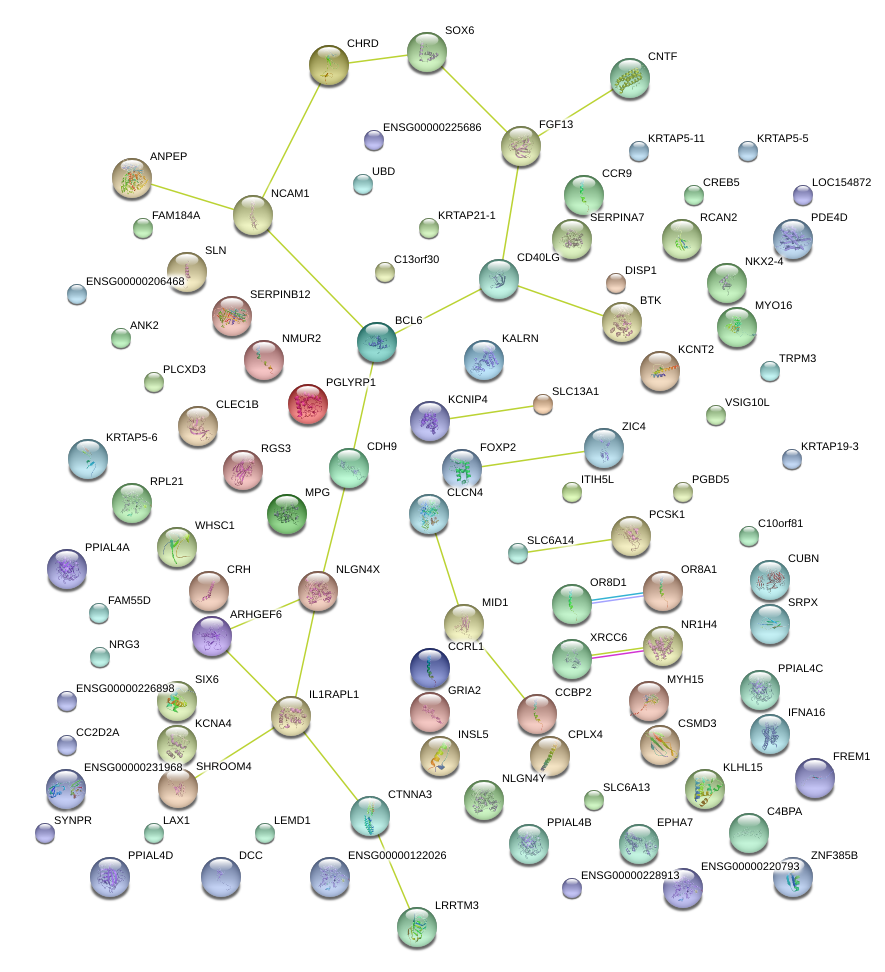
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chrX_-_50403412
|
2.648
|
|
SHROOM4
|
shroom family member 4
|
|
chr20_-_21326046
|
1.993
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chrX_-_54841397
|
1.928
|
NM_198510
|
ITIH5L
|
inter-alpha (globulin) inhibitor H5-like
|
|
chr11_-_16380967
|
1.855
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chrX_+_28515479
|
1.838
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr6_-_94185780
|
1.737
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr7_+_113842407
|
1.720
|
|
FOXP2
|
forkhead box P2
|
|
chr18_-_55136673
|
1.649
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr6_-_119512035
|
1.638
|
NM_001100411
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr9_-_72673753
|
1.637
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr2_-_180319013
|
1.458
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chrX_+_135558001
|
1.380
|
NM_000074
|
CD40LG
|
CD40 ligand
|
|
chr3_-_148604760
|
1.355
|
NM_001168378
|
ZIC4
|
Zic family member 4
|
|
chrX_-_105169370
|
1.344
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr5_-_95794416
|
1.262
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr18_+_59374372
|
1.232
|
NM_080474
|
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12
|
|
chrX_-_137649180
|
1.212
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr7_+_113842470
|
1.204
|
|
FOXP2
|
forkhead box P2
|
|
chr1_-_194844061
|
1.187
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr22_+_40372957
|
1.145
|
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6
|
|
chrX_-_6156655
|
1.139
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chrX_+_115481848
|
1.122
|
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr4_-_20594645
|
1.112
|
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chrX_+_10084984
|
1.099
|
NM_001830
|
CLCN4
|
chloride channel 4
|
|
chr1_+_221168388
|
1.092
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr1_-_142559339
|
1.087
|
NM_001123068
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G
|
|
chr11_+_123945102
|
1.087
|
NM_001005194
|
OR8A1
|
olfactory receptor, family 8, subfamily A, member 1
|
|
chr13_+_108079570
|
1.078
|
NM_001198950
|
MYO16
|
myosin XVI
|
|
chrX_-_135783496
|
1.066
|
|
|
|
|
chr10_+_115501202
|
1.064
|
NM_001193434
NM_001193435
NM_024889
|
C10orf81
|
chromosome 10 open reading frame 81
|
|
chr1_-_143075560
|
1.059
|
NM_178230
NM_001143883
NM_001135789
|
PPIAL4A
PPIAL4G
PPIAL4B
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
peptidylprolyl isomerase A (cyclophilin A)-like 4B
peptidylprolyl isomerase A (cyclophilin A)-like 4C
|
|
chr10_-_17211795
|
1.055
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr9_-_14769530
|
1.044
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chrX_+_115481774
|
1.042
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr7_+_20008870
|
1.012
|
|
RPL21
|
ribosomal protein L21
|
|
chr1_-_67039518
|
0.991
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr10_+_83627422
|
0.975
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chrX_-_23955223
|
0.966
|
NM_030624
|
KLHL15
|
kelch-like 15 (Drosophila)
|
|
chr4_+_114190233
|
0.953
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_-_188935363
|
0.952
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr3_+_185580554
|
0.941
|
NM_003741
|
CHRD
|
chordin
|
|
chr13_+_42253685
|
0.941
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr5_-_27074407
|
0.938
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr7_+_113842284
|
0.934
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr11_+_58146721
|
0.933
|
NM_000614
|
CNTF
|
ciliary neurotrophic factor
|
|
chr3_+_63403792
|
0.919
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr11_-_113971681
|
0.900
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr1_-_203657803
|
0.890
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr1_+_205344200
|
0.861
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chr4_+_158361185
|
0.854
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr3_+_125296217
|
0.851
|
NM_001024660
NM_003947
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr14_+_60045621
|
0.851
|
NM_007374
|
SIX6
|
SIX homeobox 6
|
|
chr15_-_88150823
|
0.843
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr9_+_115303527
|
0.828
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_-_21207303
|
0.827
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr18_-_21184824
|
0.826
|
|
|
|
|
chr3_+_45902999
|
0.819
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr21_-_30786145
|
0.811
|
NM_181609
|
KRTAP19-3
|
keratin associated protein 19-3
|
|
chr11_+_112337491
|
0.807
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr5_-_41546384
|
0.806
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr1_+_147819626
|
0.802
|
NM_178230
NM_001135789
|
PPIAL4A
PPIAL4G
PPIAL4C
|
peptidylprolyl isomerase A (cyclophilin A)-like 4A
peptidylprolyl isomerase A (cyclophilin A)-like 4G
peptidylprolyl isomerase A (cyclophilin A)-like 4C
|
|
chr4_+_114433551
|
0.793
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chrX_-_135577102
|
0.778
|
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr1_-_228580013
|
0.775
|
NM_024554
|
PGBD5
|
piggyBac transposable element derived 5
|
|
chr7_+_142053693
|
0.762
|
NM_001190487
|
MTRNR2L6
|
MT-RNR2-like 6
|
|
chr7_+_28418668
|
0.762
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_-_124217978
|
0.754
|
NM_001024603
|
LOC154872
|
hypothetical protein LOC154872
|
|
chr7_-_122627235
|
0.744
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr11_-_29995063
|
0.732
|
NM_002233
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4
|
|
chr5_-_151764857
|
0.731
|
NM_020167
|
NMUR2
|
neuromedin U receptor 2
|
|
chr11_-_123685871
|
0.727
|
NM_001002917
|
OR8D1
|
olfactory receptor, family 8, subfamily D, member 1
|
|
chr19_-_56537189
|
0.722
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr3_-_109730858
|
0.721
|
NM_014981
|
MYH15
|
myosin, heavy chain 15
|
|
chr4_+_15080586
|
0.719
|
NM_001080522
NM_001164720
NM_020785
|
CC2D2A
|
coiled-coil and C2 domain containing 2A
|
|
chr6_-_46401174
|
0.718
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr12_-_10042956
|
0.717
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chrX_-_37916138
|
0.717
|
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr1_-_194844299
|
0.715
|
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr10_+_68355797
|
0.713
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr11_+_112337199
|
0.701
|
NM_000615
NM_001076682
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr11_-_107087943
|
0.699
|
NM_003063
|
SLN
|
sarcolipin
|
|
chrX_-_100527799
|
0.697
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr21_-_31049566
|
0.695
|
NM_181619
|
KRTAP21-1
|
keratin associated protein 21-1
|
|
chr4_+_1923629
|
0.695
|
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr8_-_67253234
|
0.692
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chrX_-_10504852
|
0.689
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr6_-_29635456
|
0.688
|
|
UBD
|
ubiquitin D
|
|
chr11_+_1675000
|
0.684
|
NM_001012416
|
KRTAP5-6
|
keratin associated protein 5-6
|
|
chr10_-_69095362
|
0.683
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr1_+_202000906
|
0.681
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr12_+_99391681
|
0.681
|
NM_005123
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr5_-_58331515
|
0.676
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_+_112337411
|
0.674
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr18_+_48120539
|
0.669
|
NM_005215
|
DCC
|
deleted in colorectal carcinoma
|
|
chr12_-_242255
|
0.666
|
NM_001190997
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr7_-_142434340
|
0.662
|
NM_001001658
|
OR9A2
|
olfactory receptor, family 9, subfamily A, member 2
|
|
chr3_-_185578621
|
0.656
|
NM_000460
NM_001177597
NM_001177598
|
THPO
|
thrombopoietin
|
|
chr18_+_41448763
|
0.655
|
NM_007163
|
SLC14A2
|
solute carrier family 14 (urea transporter), member 2
|
|
chr1_+_65503011
|
0.651
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr10_+_50177192
|
0.646
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr1_+_146910634
|
0.645
|
NM_001144032
|
PPIAL4E
|
peptidylprolyl isomerase A (cyclophilin A)-like 4E
|
|
chr5_+_156540425
|
0.639
|
NM_005546
|
ITK
|
IL2-inducible T-cell kinase
|
|
chr7_+_142590633
|
0.637
|
NM_176881
|
TAS2R39
|
taste receptor, type 2, member 39
|
|
chrX_+_37735013
|
0.637
|
NM_012274
|
CXorf27
|
chromosome X open reading frame 27
|
|
chr13_-_25687062
|
0.635
|
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr8_+_65655367
|
0.632
|
NM_152414
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr12_-_23995220
|
0.630
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr3_+_99484421
|
0.630
|
NM_001005482
|
OR5H2
|
olfactory receptor, family 5, subfamily H, member 2
|
|
chrX_+_111005934
|
0.625
|
NM_001195576
NM_001195578
|
LOC100329135
|
hypothetical LOC100329135
|
|
chr9_-_116608113
|
0.622
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr11_+_112337299
|
0.615
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chrX_-_110542040
|
0.615
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr2_+_66519949
|
0.613
|
|
MEIS1
|
Meis homeobox 1
|
|
chr5_-_88004891
|
0.613
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr19_+_43616167
|
0.609
|
NM_000540
NM_001042723
|
RYR1
|
ryanodine receptor 1 (skeletal)
|
|
chr8_+_69026895
|
0.607
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr11_+_26310253
|
0.602
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr18_-_21186107
|
0.602
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chr13_+_30404809
|
0.601
|
NM_152325
|
C13orf26
|
chromosome 13 open reading frame 26
|
|
chr3_-_125192888
|
0.594
|
NM_017578
|
ROPN1
|
rhophilin associated tail protein 1
|
|
chr11_-_27679617
|
0.592
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr4_-_116254381
|
0.590
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr6_-_89984214
|
0.588
|
NM_002042
|
GABRR1
|
gamma-aminobutyric acid (GABA) receptor, rho 1
|
|
chr1_+_203106600
|
0.588
|
|
NFASC
|
neurofascin
|
|
chrX_+_99726445
|
0.588
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr3_-_114047479
|
0.587
|
NM_001008784
|
CD200R1L
|
CD200 receptor 1-like
|
|
chr13_-_93929719
|
0.582
|
NM_001129889
NM_001922
|
DCT
|
dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2)
|
|
chr6_-_46155937
|
0.580
|
NM_001114086
|
CLIC5
|
chloride intracellular channel 5
|
|
chr18_+_59726183
|
0.578
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr21_-_38955487
|
0.578
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr1_-_92725017
|
0.574
|
NM_005263
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chrX_+_8392870
|
0.574
|
NM_001001888
|
VCX3B
VCX3A
|
variable charge, X-linked 3B
variable charge, X-linked 3A
|
|
chr6_-_46401363
|
0.567
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr12_+_6884157
|
0.561
|
NM_001135217
NM_006992
NM_201650
|
LRRC23
|
leucine rich repeat containing 23
|
|
chr6_-_29635680
|
0.561
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chrX_+_77889861
|
0.559
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr17_-_44037332
|
0.557
|
NM_018952
|
HOXB6
|
homeobox B6
|
|
chr1_+_57093030
|
0.555
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr8_-_7718769
|
0.555
|
NM_152250
NM_001040703
|
DEFB105A
DEFB105B
|
defensin, beta 105A
defensin, beta 105B
|
|
chr8_-_110762409
|
0.555
|
NM_001099745
NM_001099746
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chrX_-_24575146
|
0.553
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chrX_+_53095230
|
0.550
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chrX_-_24575375
|
0.550
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr2_-_162808123
|
0.548
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr1_+_174698929
|
0.545
|
NM_020318
NM_021936
|
PAPPA2
|
pappalysin 2
|
|
chr11_-_56166490
|
0.545
|
NM_001002925
|
OR5AP2
|
olfactory receptor, family 5, subfamily AP, member 2
|
|
chr11_-_84312085
|
0.543
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_-_152449591
|
0.542
|
NM_001081455
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chrX_-_74921704
|
0.539
|
NM_138703
|
MAGEE2
|
melanoma antigen family E, 2
|
|
chr9_-_112010233
|
0.536
|
NM_001012993
|
C9orf152
|
chromosome 9 open reading frame 152
|
|
chr3_+_35697432
|
0.535
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr15_+_30797466
|
0.535
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr6_-_101018271
|
0.533
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr3_+_99370233
|
0.532
|
NM_001005515
|
OR5H15
|
olfactory receptor, family 5, subfamily H, member 15
|
|
chr13_+_57103944
|
0.531
|
|
PCDH17
|
protocadherin 17
|
|
chr10_-_100985548
|
0.530
|
NM_001166244
NM_001166245
NM_001166246
NM_021828
|
HPSE2
|
heparanase 2
|
|
chr10_-_43383912
|
0.529
|
NM_005674
|
ZNF239
|
zinc finger protein 239
|
|
chr3_+_138966268
|
0.526
|
NM_004189
|
SOX14
|
SRY (sex determining region Y)-box 14
|
|
chr9_+_21399132
|
0.525
|
NM_002170
|
IFNA8
|
interferon, alpha 8
|
|
chr4_+_87070449
|
0.523
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr11_-_8921155
|
0.519
|
NM_020646
|
ASCL3
|
achaete-scute complex homolog 3 (Drosophila)
|
|
chr15_-_31999815
|
0.518
|
|
|
|
|
chr1_-_200182090
|
0.515
|
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chr8_+_85258056
|
0.514
|
NM_173848
|
RALYL
|
RALY RNA binding protein-like
|
|
chr13_-_47899043
|
0.514
|
NM_001162497
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr18_-_29273979
|
0.508
|
NM_001105528
|
C18orf34
|
chromosome 18 open reading frame 34
|
|
chr12_-_90029328
|
0.507
|
|
LUM
|
lumican
|
|
chr3_+_141879555
|
0.505
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr8_+_110168828
|
0.505
|
NM_003301
|
TRHR
|
thyrotropin-releasing hormone receptor
|
|
chr5_+_50714996
|
0.504
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr9_-_14388981
|
0.501
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr11_-_113971607
|
0.501
|
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr1_-_154205675
|
0.500
|
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr11_+_19328846
|
0.497
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr12_+_54172413
|
0.497
|
NM_001005519
|
OR6C68
|
olfactory receptor, family 6, subfamily C, member 68
|
|
chr8_-_102005697
|
0.495
|
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide
|
|
chr18_-_21185968
|
0.491
|
|
ZNF521
|
zinc finger protein 521
|
|
chr8_-_127032577
|
0.488
|
|
LOC100130231
|
similar to hCG1814455
|
|
chr7_+_103756339
|
0.487
|
NM_199000
|
LHFPL3
|
lipoma HMGIC fusion partner-like 3
|
|
chr7_-_75876830
|
0.486
|
NM_080744
|
SRCRB4D
|
scavenger receptor cysteine rich domain containing, group B (4 domains)
|
|
chr12_-_80676562
|
0.485
|
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr10_-_127454379
|
0.484
|
NM_147191
|
MMP21
|
matrix metallopeptidase 21
|
|
chr18_-_63333487
|
0.483
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr4_+_81337663
|
0.482
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr11_+_110631916
|
0.482
|
NM_198498
|
C11orf53
|
chromosome 11 open reading frame 53
|
|
chr18_+_30652223
|
0.481
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr5_-_50714836
|
0.480
|
|
LOC642366
|
hypothetical LOC642366
|
|
chr12_+_99712504
|
0.480
|
NM_178826
|
ANO4
|
anoctamin 4
|
|
chr12_-_48246488
|
0.478
|
NM_001012300
|
MCRS1
|
microspherule protein 1
|
|
chr6_+_105511615
|
0.476
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr2_-_40593074
|
0.475
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr11_-_85103438
|
0.473
|
NM_032379
|
SYTL2
|
synaptotagmin-like 2
|
|
chr11_+_58888460
|
0.472
|
NM_001004729
|
OR5AN1
|
olfactory receptor, family 5, subfamily AN, member 1
|
|
chr13_+_57103789
|
0.472
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr12_-_119249938
|
0.472
|
NM_000928
|
PLA2G1B
|
phospholipase A2, group IB (pancreas)
|
|
chr18_+_29573152
|
0.472
|
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chrX_-_151370325
|
0.471
|
NM_000808
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr15_-_32833980
|
0.471
|
NM_020660
|
GJD2
|
gap junction protein, delta 2, 36kDa
|
|
chrX_-_123925343
|
0.471
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1(Drosophila)
|