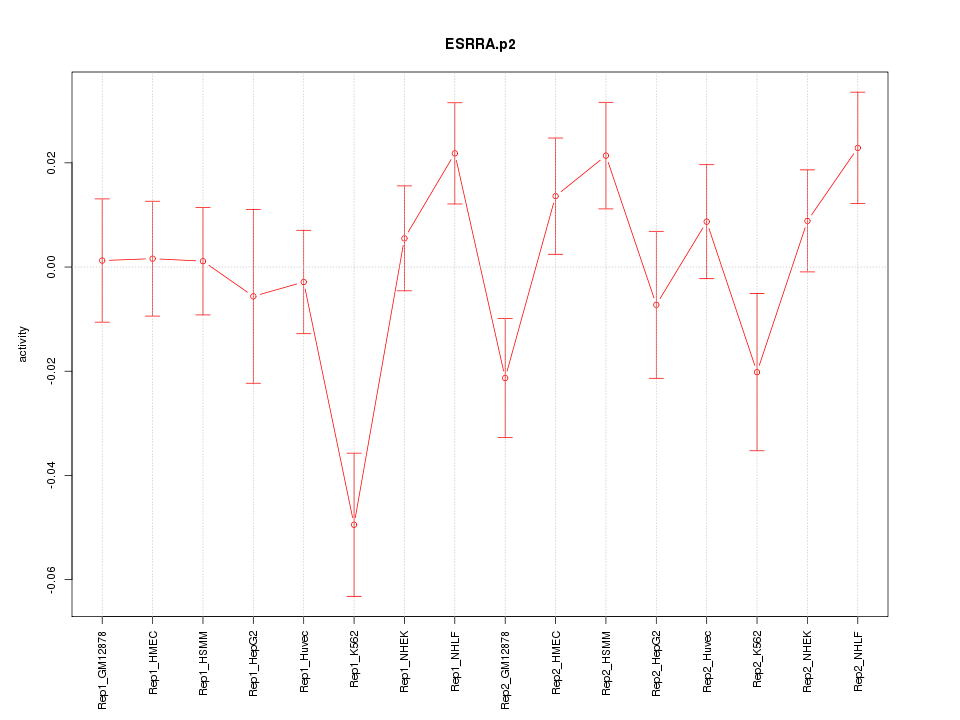
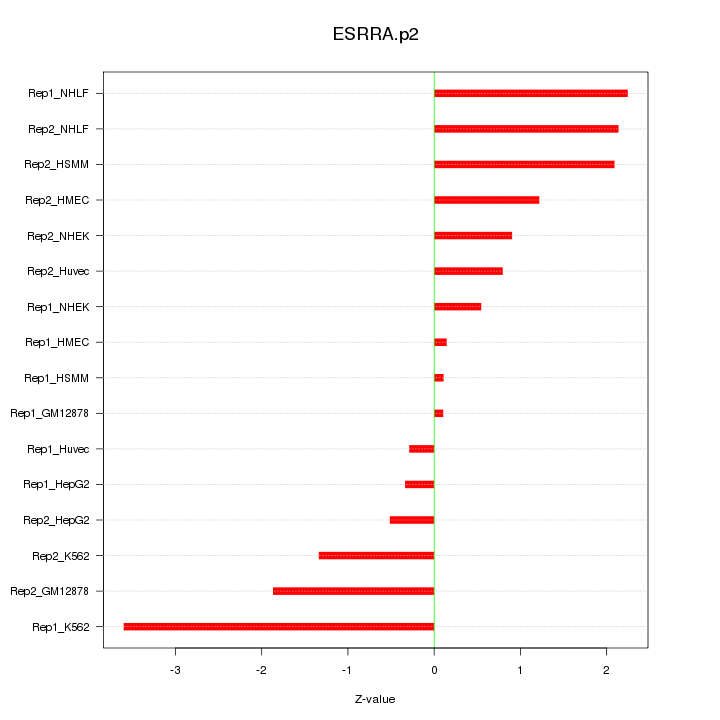
|
chr19_+_17923110
|
2.184
|
NM_002248
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1
|
|
chr9_+_17568952
|
2.131
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr5_-_148014282
|
2.115
|
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr19_+_540849
|
2.023
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr5_+_149549712
|
1.948
|
NM_014228
|
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7
|
|
chr17_-_70367479
|
1.898
|
NM_000835
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr5_-_1647617
|
1.876
|
|
SDHAP3
|
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3
|
|
chr11_+_65815940
|
1.875
|
NM_153266
|
TMEM151A
|
transmembrane protein 151A
|
|
chr22_-_29290875
|
1.860
|
NM_004861
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1
|
|
chr15_+_41672543
|
1.833
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr22_-_28041474
|
1.808
|
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr22_-_28041650
|
1.722
|
NM_001007279
NM_006477
|
RASL10A
|
RAS-like, family 10, member A
|
|
chr11_+_65815911
|
1.714
|
|
TMEM151A
|
transmembrane protein 151A
|
|
chr14_-_47213906
|
1.568
|
NM_001113498
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr19_-_4486202
|
1.520
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr20_+_36867708
|
1.482
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr9_-_21340885
|
1.474
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chrX_-_1470862
|
1.464
|
NM_001636
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6
|
|
chr10_+_44726635
|
1.458
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chr14_-_20572783
|
1.415
|
NM_001012264
|
RNASE13
|
ribonuclease, RNase A family, 13 (non-active)
|
|
chr5_-_63293301
|
1.405
|
NM_000524
|
HTR1A
|
5-hydroxytryptamine (serotonin) receptor 1A
|
|
chr11_-_64268841
|
1.351
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr12_+_20413445
|
1.332
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr11_-_64268126
|
1.321
|
NM_001098671
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr4_-_21308415
|
1.234
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr22_-_42589543
|
1.214
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr17_-_70430839
|
1.212
|
NM_173477
|
USH1G
|
Usher syndrome 1G (autosomal recessive)
|
|
chr14_-_103058865
|
1.204
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr3_-_27738788
|
1.199
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr22_+_23221114
|
1.182
|
NM_016327
|
UPB1
|
ureidopropionase, beta
|
|
chr3_+_142253432
|
1.163
|
NM_080862
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
|
chr12_-_116021473
|
1.155
|
NM_001168325
NM_017899
|
TESC
|
tescalcin
|
|
chr11_-_132318080
|
1.102
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr14_+_69021149
|
1.093
|
NM_001161498
|
UPF0639
|
UPF0639 protein
|
|
chr1_+_29086206
|
1.092
|
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr10_+_105334704
|
1.091
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr12_+_56299959
|
1.071
|
NM_133489
|
SLC26A10
|
solute carrier family 26, member 10
|
|
chr11_-_132318741
|
1.063
|
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr17_+_7549138
|
1.045
|
NM_001406
|
EFNB3
|
ephrin-B3
|
|
chr5_-_148013908
|
1.045
|
NM_000870
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr3_+_134947788
|
1.032
|
|
TF
|
transferrin
|
|
chr15_+_41772375
|
1.027
|
NM_001015001
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr19_-_60360768
|
1.020
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr22_+_29848923
|
1.011
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr5_-_35266305
|
1.008
|
|
PRLR
|
prolactin receptor
|
|
chr17_+_71586857
|
1.003
|
NM_180990
|
ZACN
|
zinc activated ligand-gated ion channel
|
|
chr19_-_44518516
|
1.001
|
NM_004877
|
GMFG
|
glia maturation factor, gamma
|
|
chr3_-_180805104
|
0.933
|
NM_020409
NM_177988
|
MRPL47
|
mitochondrial ribosomal protein L47
|
|
chr12_-_642786
|
0.927
|
NM_016533
|
NINJ2
|
ninjurin 2
|
|
chr1_+_29086156
|
0.914
|
NM_001166005
NM_001166007
NM_004437
NM_203342
NM_203343
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr20_-_61600834
|
0.907
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr3_+_134947940
|
0.905
|
|
TF
|
transferrin
|
|
chr5_-_35266550
|
0.884
|
NM_000949
|
PRLR
|
prolactin receptor
|
|
chr3_+_134947894
|
0.883
|
|
TF
|
transferrin
|
|
chr12_-_52980914
|
0.872
|
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr12_-_52980984
|
0.869
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr11_-_84311867
|
0.867
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_-_10724715
|
0.863
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr5_+_149550003
|
0.863
|
|
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7
|
|
chr1_-_152431175
|
0.858
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr18_-_55136673
|
0.845
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr18_-_5533967
|
0.827
|
NM_012307
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3
|
|
chrX_-_132377087
|
0.813
|
|
GPC4
|
glypican 4
|
|
chr7_-_100630912
|
0.811
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr17_-_34014390
|
0.806
|
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr11_+_268569
|
0.796
|
NM_138329
|
NLRP6
|
NLR family, pyrin domain containing 6
|
|
chr11_-_45264193
|
0.796
|
NM_020826
|
SYT13
|
synaptotagmin XIII
|
|
chr10_-_103525646
|
0.795
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chrX_+_48505097
|
0.793
|
NM_001080489
|
GLOD5
|
glyoxalase domain containing 5
|
|
chr15_-_76210611
|
0.791
|
NM_006383
|
CIB2
|
calcium and integrin binding family member 2
|
|
chr3_+_9720509
|
0.787
|
NM_153635
|
CPNE9
|
copine family member IX
|
|
chr4_-_44145266
|
0.765
|
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr20_-_43767069
|
0.765
|
NM_172006
NM_172131
|
WFDC10B
|
WAP four-disulfide core domain 10B
|
|
chr12_+_76749199
|
0.765
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr22_+_35586950
|
0.758
|
NM_000631
NM_013416
|
NCF4
|
neutrophil cytosolic factor 4, 40kDa
|
|
chr12_-_107257205
|
0.752
|
NM_001142343
NM_001142344
NM_004072
|
CMKLR1
|
chemokine-like receptor 1
|
|
chr3_+_134947573
|
0.750
|
NM_001063
|
TF
|
transferrin
|
|
chr8_-_61356436
|
0.732
|
NM_004056
|
CA8
|
carbonic anhydrase VIII
|
|
chr8_+_120289735
|
0.725
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr5_+_176717553
|
0.723
|
|
|
|
|
chr20_-_24987615
|
0.719
|
NM_032501
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1
|
|
chr20_+_41621119
|
0.701
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr18_+_42751510
|
0.700
|
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr1_+_234916376
|
0.700
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr20_-_44713466
|
0.699
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr2_+_219453487
|
0.697
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr7_+_45580704
|
0.693
|
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr3_-_152449591
|
0.692
|
NM_001081455
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14
|
|
chr1_+_157824239
|
0.681
|
NM_001639
|
APCS
|
amyloid P component, serum
|
|
chr19_+_50693570
|
0.677
|
NM_001080401
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative)
|
|
chr15_+_76644882
|
0.676
|
NM_000745
|
CHRNA5
|
cholinergic receptor, nicotinic, alpha 5
|
|
chr2_+_74594093
|
0.652
|
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr4_+_44375165
|
0.645
|
NM_021927
|
GUF1
|
GUF1 GTPase homolog (S. cerevisiae)
|
|
chr10_-_97190885
|
0.645
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr8_+_36760999
|
0.638
|
NM_001031836
|
KCNU1
|
potassium channel, subfamily U, member 1
|
|
chr14_+_87560646
|
0.636
|
|
LOC283587
|
hypothetical protein LOC283587
|
|
chr19_+_19183763
|
0.632
|
NM_004386
|
NCAN
|
neurocan
|
|
chr19_-_44518465
|
0.625
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr5_+_176807931
|
0.619
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr20_+_41621079
|
0.617
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr2_-_31293882
|
0.617
|
NM_001145122
|
CAPN14
|
calpain 14
|
|
chr7_+_45580606
|
0.612
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr14_+_99220349
|
0.611
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr13_+_32488477
|
0.596
|
NM_004795
|
KL
|
klotho
|
|
chr12_-_16652202
|
0.596
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr14_+_41146513
|
0.588
|
NM_152447
|
LRFN5
|
leucine rich repeat and fibronectin type III domain containing 5
|
|
chr14_-_74400288
|
0.587
|
NM_001080408
|
PROX2
|
prospero homeobox 2
|
|
chr10_+_68355797
|
0.585
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chrX_+_147389825
|
0.585
|
NM_001169122
NM_001169123
NM_001169124
NM_001169125
NM_002025
|
AFF2
|
AF4/FMR2 family, member 2
|
|
chr11_-_64582568
|
0.579
|
NM_005468
|
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1
|
|
chr8_+_100025806
|
0.573
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr14_+_36200867
|
0.571
|
|
PAX9
|
paired box 9
|
|
chr13_-_51483473
|
0.566
|
NM_000053
NM_001005918
|
ATP7B
|
ATPase, Cu++ transporting, beta polypeptide
|
|
chr1_+_32489426
|
0.549
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr3_+_51716120
|
0.547
|
NM_000839
NM_001130063
|
GRM2
|
glutamate receptor, metabotropic 2
|
|
chr14_-_69616616
|
0.546
|
NM_182936
NM_001130417
|
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3
|
|
chr14_-_73296643
|
0.539
|
NM_194278
|
C14orf43
|
chromosome 14 open reading frame 43
|
|
chr8_-_100975417
|
0.533
|
NM_004374
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr14_+_36200815
|
0.529
|
|
PAX9
|
paired box 9
|
|
chr14_+_92459244
|
0.527
|
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr1_-_172153024
|
0.526
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr12_-_3732478
|
0.524
|
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr1_-_19089971
|
0.520
|
NM_001161504
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr17_+_37247568
|
0.518
|
NM_152467
|
KLHL10
|
kelch-like 10 (Drosophila)
|
|
chr2_-_154043403
|
0.510
|
NM_019845
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate
|
|
chr8_-_100975086
|
0.503
|
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr14_-_103058568
|
0.503
|
|
CKB
|
creatine kinase, brain
|
|
chr22_+_40702709
|
0.503
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr7_+_37926687
|
0.496
|
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr19_+_55614006
|
0.493
|
NM_003121
|
SPIB
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr11_+_43920385
|
0.490
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr3_-_134863316
|
0.490
|
NM_007027
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1
|
|
chr2_+_104838177
|
0.488
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr5_+_271428
|
0.484
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr22_-_29197944
|
0.483
|
NM_174975
|
SEC14L3
|
SEC14-like 3 (S. cerevisiae)
|
|
chr2_+_222997456
|
0.481
|
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr1_-_172153087
|
0.481
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr20_-_1922393
|
0.480
|
NM_001190900
|
PDYN
|
prodynorphin
|
|
chr2_+_222997529
|
0.478
|
NM_152386
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr17_+_45778381
|
0.477
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr22_-_22440054
|
0.473
|
NM_213720
|
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr15_+_38520594
|
0.473
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr5_-_45731976
|
0.470
|
NM_021072
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1
|
|
chr20_+_1132097
|
0.467
|
NM_001009612
|
C20orf202
|
chromosome 20 open reading frame 202
|
|
chr6_+_31662454
|
0.457
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr2_+_119905812
|
0.456
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr2_-_30883775
|
0.455
|
NM_144575
|
CAPN13
|
calpain 13
|
|
chr19_-_12773591
|
0.454
|
NM_005809
NM_181738
|
PRDX2
|
peroxiredoxin 2
|
|
chr17_-_4747147
|
0.445
|
NM_000080
|
CHRNE
|
cholinergic receptor, nicotinic, epsilon
|
|
chr22_-_48866484
|
0.443
|
NM_015166
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr5_+_271441
|
0.441
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr8_+_11599119
|
0.441
|
NM_002052
|
GATA4
|
GATA binding protein 4
|
|
chr22_+_30480848
|
0.438
|
NM_001136029
|
DEPDC5
|
DEP domain containing 5
|
|
chr3_-_148605996
|
0.434
|
NM_001168379
|
ZIC4
|
Zic family member 4
|
|
chr20_+_3000265
|
0.422
|
NM_000915
|
OXT
|
oxytocin, prepropeptide
|
|
chr9_-_99994704
|
0.417
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr12_+_119647953
|
0.415
|
NM_000017
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain
|
|
chr22_-_48865907
|
0.406
|
NM_139202
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr5_-_150517528
|
0.403
|
NM_001155
|
ANXA6
|
annexin A6
|
|
chr18_-_42590989
|
0.398
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr5_+_271447
|
0.392
|
|
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp)
|
|
chr3_+_130451160
|
0.392
|
|
COPG
|
coatomer protein complex, subunit gamma
|
|
chr4_+_7245218
|
0.392
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr6_+_44418374
|
0.391
|
NM_145026
|
SPATS1
|
spermatogenesis associated, serine-rich 1
|
|
chrX_-_53002900
|
0.390
|
NM_014138
|
FAM156A
|
family with sequence similarity 156, member A
|
|
chr9_-_88064338
|
0.386
|
NM_001010907
|
C9orf153
|
chromosome 9 open reading frame 153
|
|
chr22_-_48865980
|
0.383
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr2_-_86871459
|
0.381
|
|
CD8A
|
CD8a molecule
|
|
chr4_-_40212670
|
0.379
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr22_-_22646602
|
0.375
|
NM_001084392
|
DDT
|
D-dopachrome tautomerase
|
|
chr8_+_55533047
|
0.374
|
NM_022454
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr11_+_22652957
|
0.373
|
|
GAS2
|
growth arrest-specific 2
|
|
chr2_+_218843358
|
0.370
|
NM_001077399
NM_015488
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr1_-_199259447
|
0.370
|
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr3_-_50624085
|
0.368
|
|
CISH
|
cytokine inducible SH2-containing protein
|
|
chr7_+_138859213
|
0.368
|
NM_001080511
|
CLEC2L
|
C-type lectin domain family 2, member L
|
|
chr4_+_36923084
|
0.367
|
NM_001144990
|
KIAA1239
|
KIAA1239
|
|
chr8_-_93176819
|
0.367
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_-_32540818
|
0.367
|
|
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase
|
|
chr20_+_3724905
|
0.365
|
|
CDC25B
|
cell division cycle 25 homolog B (S. pombe)
|
|
chr3_-_132704339
|
0.364
|
NM_007208
|
MRPL3
|
mitochondrial ribosomal protein L3
|
|
chrX_-_21686079
|
0.363
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr5_-_150707281
|
0.361
|
NM_181776
|
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2
|
|
chr8_-_100975041
|
0.360
|
|
COX6C
|
cytochrome c oxidase subunit VIc
|
|
chr5_-_150517499
|
0.356
|
|
ANXA6
|
annexin A6
|
|
chr4_+_24406336
|
0.355
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr11_+_82545784
|
0.349
|
NM_015885
|
PCF11
|
PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae)
|
|
chr11_+_65140207
|
0.347
|
NM_032223
|
PCNXL3
|
pecanex-like 3 (Drosophila)
|
|
chr1_+_55237193
|
0.346
|
NM_057176
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin)
|
|
chr5_-_137938904
|
0.343
|
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin)
|
|
chr12_-_16650697
|
0.343
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_-_86942504
|
0.343
|
NM_001178100
NM_004931
NM_172101
NM_172102
NM_172213
|
CD8B
|
CD8b molecule
|
|
chr22_-_48865864
|
0.342
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr19_+_54621499
|
0.341
|
NM_001195256
|
LOC645971
|
hypothetical protein LOC645971
|
|
chrX_-_132376660
|
0.339
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr1_+_119759076
|
0.338
|
NM_001166120
|
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2
|
|
chr9_-_116190038
|
0.337
|
|
AKNA
|
AT-hook transcription factor
|
|
chr2_+_6935246
|
0.336
|
NM_080657
|
RSAD2
|
radical S-adenosyl methionine domain containing 2
|
|
chr19_+_50863341
|
0.334
|
NM_000164
|
GIPR
|
gastric inhibitory polypeptide receptor
|
|
chr3_+_182900081
|
0.334
|
|
SOX2OT
|
SOX2 overlapping transcript (non-protein coding)
|